Enabling the future of Cancer Precision Medicine and Research via Rapid and High-Quality Amplicon Sequencing
According to WHO, 9.6 million people worldwide are estimated to have died from cancer in 2018. On the molecular level, cancer is a disease originated from malignant DNA mutations and subsequent changes of gene expression patterns that trigger uncontrolled cell division. In fact, two cancers with the same histologic origin may be different in terms of disease progression and treatment response. These differences result from unique genetic content of each cancer. Therefore, understanding the genetic makeup of cancer specimens is critical for elucidating the underlying biology.
Next-Generation Sequencing (NGS), especially targeted sequencing, has allowed for the genetic profiling of individual patient tumors, enabling doctors to tailor therapies based on their patient’s specific cancer mutational profile. Hybrid capture and multiplex PCR are the two main targeted sequencing methods. Due to its sensitivity and workflow issue, hybrid capture-based sequencing can be less effective in dealing with samples containing either low amount of DNA or low frequency cancer mutations among normal DNA background. Compared to hybrid capture, PCR amplicon sequencing could be a better choice mainly because it is highly sensitive, cost-effective and easy to implement in routine laboratory use.
CleanPlex DNA and RNA Amplicon Sequencing Technologies – Simple, Affordable Solution for the future of Cancer Precision Medicine and Research
CleanPlex DNA Technology is a patented, scalable and ultra-sensitive multiplex PCR-based targeted DNA sequencing technology for next-generation sequencing (NGS). CleanPlex DNA workflow (Figure 1) starts with extracted DNA from fresh frozen tissue samples, FFPE tissues, cancer liquid biopsy samples, etc. The streamlined protocol can be completed in just 3 hours. Scientists or clinicians have the option to add molecular barcodes (unique molecular identifiers – UMIs) into amplicon libraries for more accurate detection of low frequency variants via CleanPlex UMI Technology.
To meet the needs of different customers, Paragon Genomics has developed a series of ready-to-use cancer panels including CleanPlex OncoZoom Cancer Hotspot Panel, CleanPlex Tumor Mutational Burden 500 Gene Panel, CleanPlex BRCA1 & BRCA2 Panel, CleanPlex® Hereditary Cancer Panel, and CleanPlex TP53 Panel.
In addition to ready-to-use panels, Paragon Genomics also offers CleanPlex Custom NGS Panels powered by CleanPlex Technology and ParagonDesigner (advanced multiplex PCR primer design software). Researchers and clinicians pick targets or genes that they would like to interrogate, and our NGS bioinformatics experts design and deliver the custom assays in complete kit format (input to sequencing-ready NGS libraries) in just 3 to 4 weeks. Our bioinformatics design team has experience in designing NGS panels for the solid tumor cancer types such as lung cancer, breast cancer, ovarian cancer, colorectal cancer, prostate cancer, cervical cancer, melanoma, pancreatic cancer, thyroid cancer, etc. and blood cancer types such as leukemia, lymphoma, and myeloma. Our team has also worked on custom panels related to homologous recombination deficiency (HRD), tumor mutational burden (TMB), MET Exon 14 Skipping, etc.
Figure 1. CleanPlex DNA Target Enrichment and Library Preparation Workflow
Figure 2. CleanPlex RNA Target Enrichment and Library Preparation Workflow
CleanPlex RNA Technology is an ultra-high multiplex PCR-based target sequencing solution for RNA samples. CleanPlex RNA workflow (Figure 2) adds an additional proprietary reverse transcription step in front of CleanPlex DNA workflow so as to convert RNA to cDNA for downstream multiplex PCR amplification. By doing so, CleanPlex RNA inherits all the advantages and features of CleanPlex DNA technologies.
CleanPlex RNA applications include single cell RNA analysis, gene expression analysis, microRNA, cancer fusion gene detection etc. Paragon Genomics has launched several CleanPlex RNA-based ready-to-use cancer fusion panels. We also accept CleanPlex RNA Custom Panel requests from researchers and clinicians.
CleanPlex Research Applications in Oncology

Prof. Elliot Stieglitz‘s Lab at the UCSF Benioff Children’s Hospital developed a 25-gene custom NGS assay powered by Paragon Genomics’ CleanPlex technology to retrospectively measure the molecular burden of juvenile myelomonocytic leukemia (JMML) patients at the time of initial diagnosis and posttherapy.
When we decided to choose a target enrichment platform, we had several choices and I am delighted that we chose Paragon Genomics. Paragon Genomics has enabled us to move our sequencing in-house with an affordable and easy-to-use product. They assisted us every step along the way from choosing the amplicons, to guiding us through library preparation to assisting with analysis. I strongly recommend their CleanPlex amplicon-based targeted sequencing approach and could not be happier with their product.
There are a wide range of instruments and methods for capturing, enriching, and enumerating circulating tumor cells (CTCs). Once isolated, CTCs can be studied using next-generation sequencing (NGS) to obtain their genomic profiles. Targeted sequencing of CTCs using amplicon-based NGS panels is an ideal approach because the workflow is simple to implement and requires little amount of input DNA.
In collaboration with the expert teams at RareCyte and Mayo Clinic, CTCs were visually identified using RareCyte’s CyteFinder® Instrument and were mechanically retrieved with RareCyte’s CytePicker® Module for genomics analysis. Cells were lysed in a PCR-compatible lysis buffer without whole genome amplification, and the lysate were used to prepare targeted NGS libraries using the CleanPlex OncoZoom Cancer Hotspot Panel. This approach resulted in libraries with excellent and consistent coverage and low error frequencies (Figure 3), enabling efficient and accurate assessment of somatic mutations in CTCs.
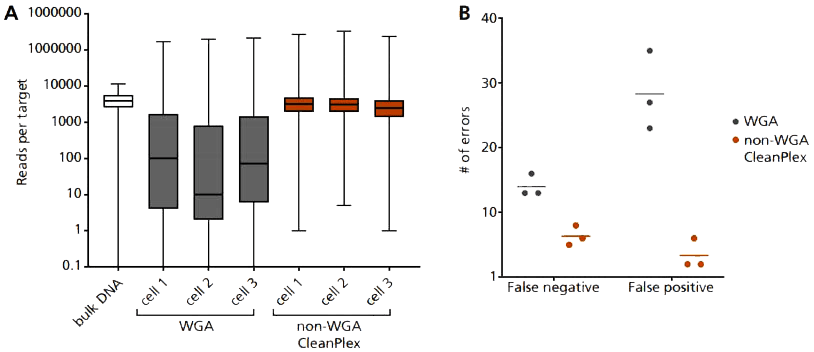
Figure 3. Single cell targeted DNA sequencing with CleanPlex OncoZoom. Single cell lysate is input as template into Paragon’s CleanPlex OncoZoom, with modified primer concentration, PCR cycle number, and clean-up steps to compensate for low input DNA concentration. Using this non-WGA method vastly improves: (A) Coverage uniformity, and incidence of (B) false negative and false positive errors, when compared to single cell WGA products.


Tumor mutational burden (TMB) is gaining tremendous interest in recent years as a biomarker particularly for predicting response to immuno-oncology (IO) therapies. TMB is measured as the total number of mutations per megabase within the coding region of a tumor genome. Patients with high TMB have been found to have more favorable response to checkpoint inhibitors, such as atezolizumab and pembrolizumab, than those with lower TMB.
Paragon Genomics has developed a high-performance CleanPlex TMB Panel that contains over 500 genes covered by approximately 27,000 amplicons. One custom version of the panel was tested by Berry Oncology (subsidiary of Berry Genomics in China) to measure TMB in solid tumor FFPE samples. For each sample, the TMB was calculated, the types of mutations were documented, and the top mutated genes were identified. They compared the mutations identified using the TMB panel to those identified with WES for the same 103 samples and found a high degree of overlap between the two methods. They also found a high degree of correlation when they compared the TMB calculated using the two methods.
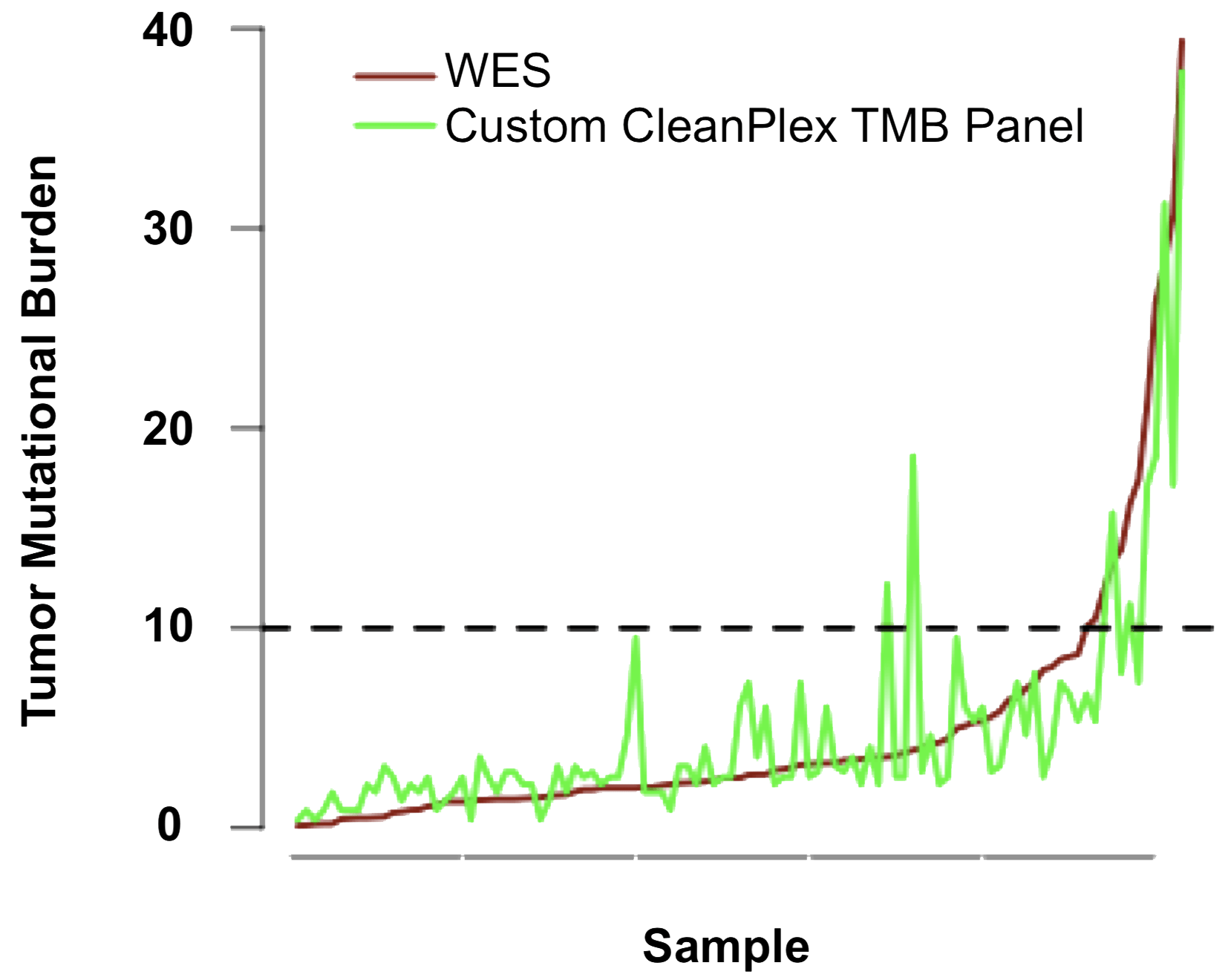
Figure 4. CleanPlex Custom TMB Panel’s High TMB Correlation with Whole Exome Sequencing Data
Request a Free Consultation for Precision Oncology Research or Testing
High-Performance Amplicon Sequencing Solution for Cancer Research and Precision Medicine
At Paragon Genomics, we take your privacy and data very seriously. You can review our Privacy Policy online or download the PDF.