NGS Amplicon Sequencing
What is Amplicon Sequencing?
Amplicon sequencing is a method of targeted next generation sequencing (NGS) that enables researchers to analyze genetic variations in specific genomic regions using polymerase chain reaction (PCR) primers designed to amplify a region (regions) of interest in a genome. Amplicons are DNA fragments of a PCR reaction and the term is often used interchangeably with “PCR product”. By creating amplicons and thus increasing the number of copies or a certain DNA region of interest, you allow for higher signals during sequencing, which in turn allows for more confident sequencing results.
Amplicon sequencing is typically used for variant detection and single-base characterization of complex samples. This approach can detect rare somatic mutations, hot-spot mutations, insertions, deletions, copy number variations, gene fusions, and single- nucleotide polymorphisms (SNPs). Because of the sensitivity and versatility of the approach, amplicon sequencing has reached and established itself in a range of disciplines, including precision oncology, inherited disease testing, pathogen detection, public health (like the recent SARS COV-2 pandemic), agricultural genomics, etc.
Amplicon Sequencing vs Hybrid Capture
The two most commonly used techniques for NGS targeted sequencing are multiplex PCR-based (amplicon sequencing) and hybrid capture-based target enrichment.
In general, amplicon sequencing method is faster, easier and cheaper than hybridization-based alternatives and so more suited for quick gene panel testing and production-scale applications such as clinical diagnostics and industrial genomic screening. Amplicon sequencing also allows for dealing with low DNA input which is difficult for hybrid captured based technology. For example, when dealing with viral detection, amplicon sequencing has a clear advantage against hybrid capture based methods in terms of detection sensitivity and DNA/RNA input requirements.
On the other hand, hybrid capture-based method can interrogate very large target region up to human whole exome and is generally better at detecting structural variations such as novel gene fusions, etc., so it is more suitable for research and discovery projects. However, capture-based methods suffer from low on-target rate for smaller panels due to the inherent lower specificity and sensitivity of hybridization probes. Due to the above reasons, scientists and clinicians tend to employ multiplex PCR for small-mid sized (less than 1Mb target region) panels for the detection of single nucleotide polymorphisms (SNPs) and/or small insertions/deletions and hybrid capture for large (0.5Mb – 50Mb target region) panels for the detection of SNPs, fusion genes, copy number variations (CNV), etc.
Below is a comparison of amplicon sequencing and hybrid capture-based target enrichment workflows.
Amplicon Sequencing Library Preparation Workflow
A Typical Hybrid Capture Target Enrichment workflow
Below is a summary of the pros and cons of both methods.
Note: Some comments on amplicon sequencing methods do not apply to Paragon Genomics’ CleanPlex technology as it overcomes many of the key drawbacks of traditional amplicon-based methods. This will be discussed in the next section.
CleanPlex® – an ultra-scalable and ultra-sensitive amplicon sequencing method that bridges the gap between traditional amplicon sequencing and hybrid capture sequencing
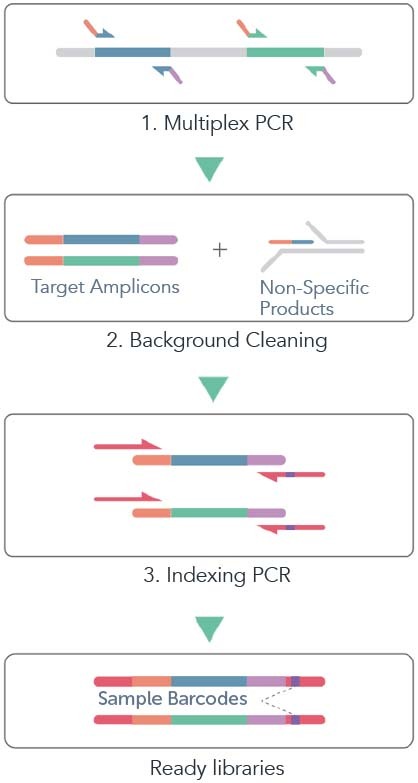
Overcoming Background Noise
First, the target regions of a genome or DNA sample are amplified by well-designed multiplex PCR primers with overhanging tails being partial adaptor sequences compatible with corresponding DNA sequencers, resulting in both target amplicons and non-specific PCR products including primer dimers.
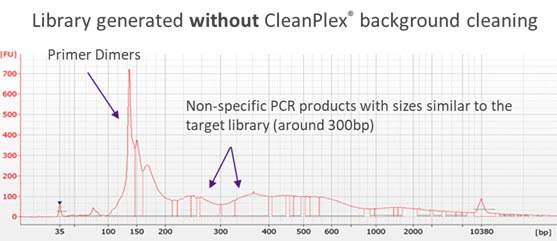
Traditionally, when the panel size is large (e.g. more than 2,000 amplicons in a single pool), the non-specific PCR products can be overwhelming and significantly affect the downstream steps if there is no measure to remove them. Some amplicon-based methods utilize bead purification and size selection to remove smaller DNA fragments such as primer dimers. However, some complicated non-specific PCR products with sizes similar to the lengths of target amplicons and its resulting libraries can be difficult to remove using just size selection. The following Bioanalyzer trace shows significant background noise around a target library of 300bp.
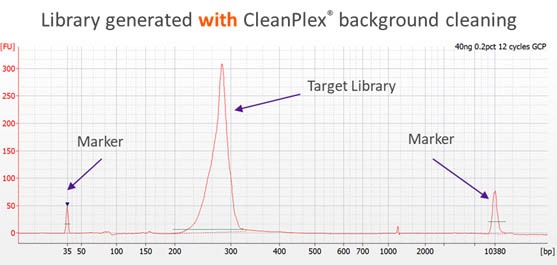
CleanPlex overcomes this drawback with an innovative and patented enzymatic / chemical background cleaning step that removes non-specific PCR products including both primer dimers and more complicated and longer nonspecific PCR artifacts, resulting in very pure target libraries. The following Bioanalyzer trace shows the effect of CleanPlex background cleaning technology.
Subsequently, sample barcodes (for sample pooling purpose) are added by an indexing PCR step to get sequencing-ready libraries. The whole workflow only takes 3 hours and minimal hands-on time.
Overcoming Scalability or Panel Size Limitation
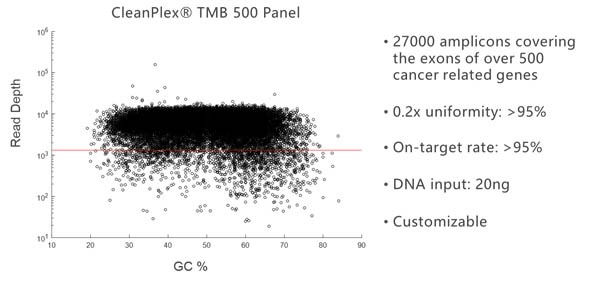
Due to its background cleaning technology, CleanPlex can easily break the panel size limit of traditional multiplex PCR and amplify more than 20,000 targets in a single panel. The following amplification plot shows the GC% of each target amplicon vs. its sequencing depth for a 27,000-amplicon panel.
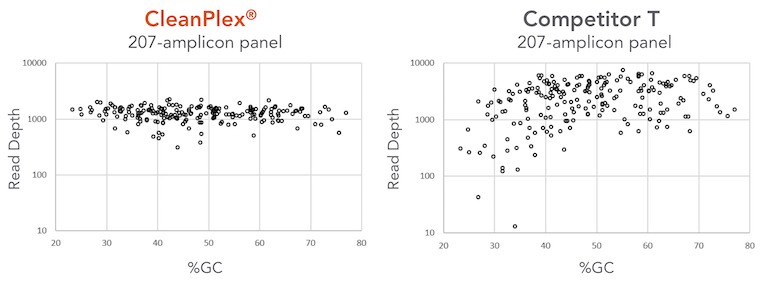
Overcoming GC Bias and Uniformity Issues
The figure below compares CleanPlex and another well-known amplicon-based method in terms of GC bias and amplification uniformity. Two panels target the same regions of a few cancer-related genes. The competitor method obviously has GC bias around low GC region and an overall low amplification uniformity across the spectrum of the panel. On the contrary, CleanPlex can amplify all target regions evenly. The result of this is that users of the competitor’s panel have to raise the average sequencing depth by 100% to achieve similar variant calling quality, therefore doubling the sequencing depth and cost.
Pushing the limit of LOD
Our collaborators RareCyte, Inc and Mayo Clinic have successfully demonstrated that CleanPlex technology can directly amplify DNA without whole genome amplification (WGA) from a single cell (only ~6 pg of DNA), specifically circulating tumor cells (CTCs) isolated from cancer patients’ blood.
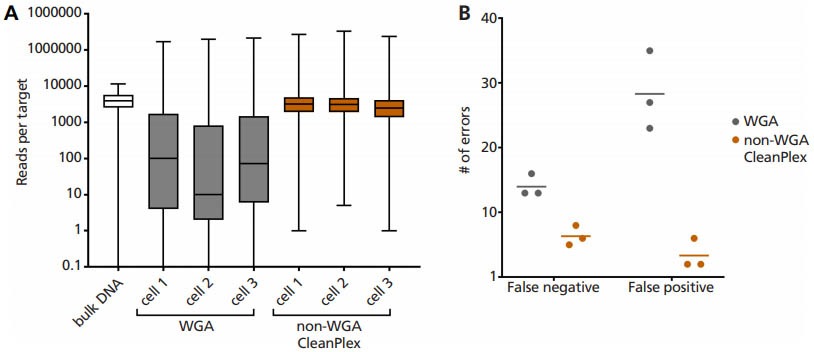
Single cell lysate is input as template into Paragon Genomics’ CleanPlex OncoZoom Panel, with modified primer concentration, PCR cycle number, and clean-up steps to compensate for low input DNA concentration. Using this non-WGA method vastly improves: (A) coverage uniformity, and incidence of (B) false negative and false positive errors, when compared to single cell WGA products.
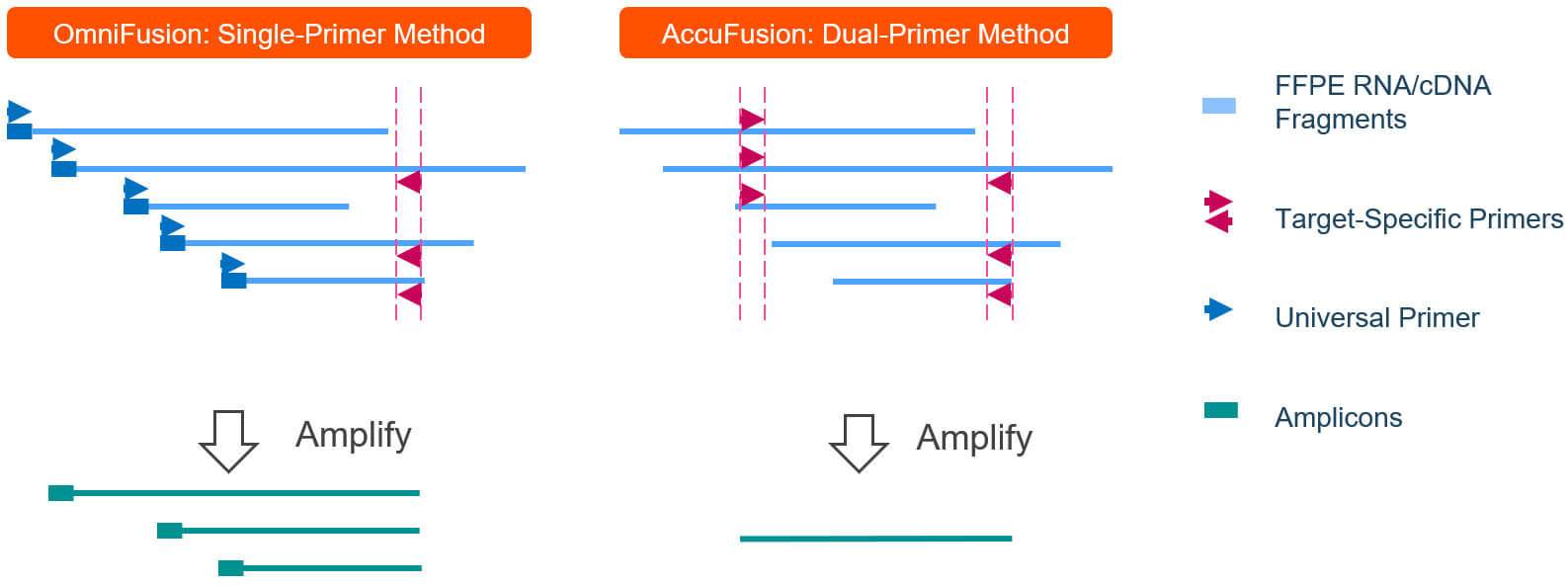
Overcoming limitation on Detecting Novel Fusion Genes
CleanPlex single primer fusion detection method – OmniFusion leverages template switching and single primer amplification to detect novel fusion genes.
In summary, CleanPlex Amplicon Sequencing Technology leverages the speed, sensitivity, workflow and cost advantages of multiplex PCR for NGS target enrichment while overcoming its key drawbacks, therefore bridging the gap between traditional amplicon-based target enrichment method and hybrid captured-based approach.
| Traditional Amplicon- or PCR-based Target Enrichment | CleanPlex Improvements |
| Pros | |
| Requires lower input of DNA (1-10 ng) | CleanPlex can directly amplify single cell DNA (6 pg) |
| Shorter and easy workflow (3-6 hours) | Advantages maintained |
| High on-target rate (>95%) | Advantages maintained |
| No special equipment required (e.g. no need of DNA fragmentation) | Advantages maintained |
| Better performance on difficult clinical samples such as FFPE tissue DNA | Advantages maintained |
| Lower reagent and consumable (pipette tips) costs | Advantages maintained |
| Cons | |
| PCR amplification bias resulting in lower assay uniformity, especially evident for large panels | Can achieve more than 95% uniformity (0.2x) even for large panels involving over 20,000 amplicons |
| Non-specific PCR background noise (e.g. primer dimer) can be high, especially evident for large panels involving over 1,000 amplicons in a single pool | Proprietary CleanPlex background cleaning technology can effectively remove background noise regardless of panel sizes |
| Difficult to design a large number of multiplex PCR primer pairs that are compatible in a single pool with minimal interaction | ParagonDesigner™ algorithm takes into account multiple primer design factors and ensures best performance for even very large panels |
| Application Limitation | |
| Novel fusion detection (cannot design primers for unknown fusion breakpoints) | CleanPlex OmniFusion single primer technology enables detection of novel fusions |
| Whole Exome Sequencing (target region can be too large and uniformity will suffer) | CleanPlex is the most scalable and uniform multiplex-PCR based technology and will be able to amplify whole exome. |
Discover more with less™
The combination of superior primer design and innovative multiplex PCR-based target enrichment and library preparation chemistry give rise to CleanPlex NGS Amplicon Panels’ ultra-high multiplexing capability, high performance, low input requirement, high sensitivity, single-tube workflow, and cost-effective amplicon sequencing. These remarkable features and benefits allow researchers and assay developers to discover more with less.
Discover More
Discover more with CleanPlex NGS Panels
• Multiplex 20,000+ amplicons per reaction
• High target design rate
• High coverage uniformity
• High on-target rate
• High sensitivity (1% LOD with 10 ng input)
Use Less
Use less input and resources to reduce costs
• Inputs as low as 10 ng
• Fast 3-hour protocol
• Simple, streamlined workflow
• Efficient use of NGS reads
Have specific requests or questions? Schedule a free consultation with our PhD-level expert scientists to learn how our best-in-class amplicon sequencing custom panel design services can help you advance your work.
View more data on CleanPlex® technology for amplicon sequencing
FAQ for Amplicon Sequencing
Amplicon sequencing is a method of targeted next generation sequencing (NGS) that enables researchers to analyze genetic variation in specific genomic regions using primers designed to target a region of interest. Amplicons are DNA fragments of a polymerase chain reaction (PCR) and the term is often used interchangeably with “PCR product”. By creating amplicons and thus increasing the number of copies or a certain DNA region of interest, you allow for higher signals during sequencing, which in turn allows for more confident sequences.
Amplicon sequencing is typically used for variant detection and single-base characterization of complex samples. This approach can detect rare somatic mutations, hot-spot mutations, insertions, deletions, copy number variations, gene fusions, and single- nucleotide polymorphisms (SNPs). Because of the sensitivity and versatility of the approach, amplicon sequencing has reached and established itself in a range of disciplines, including oncology, nutrition, public health (like the recent SARS COV-2 pandemic), microbial genomics, etc.
In amplicon sequencing, amplicons are generated by PCR or multiplex PCR, pooled and later sequenced. This highly targeted approach enables thousands of regions in the genome to be determined and amplified simultaneously. Furthermore, amplicons originating from different samples can be combined and sequenced together by adding a unique barcode or index to each individual sample, which is later used as leverage to demultiplex for downstream analysis, simplifying subsequent data interpretation and processing.
DNA stands for DeoxyriboNucleic Acid and it contains the genetic information for each organism to help them grow, function, and reproduce. The genetic code is made up of four nucleotides that organize themselves linearly. These four nucleotides are Thymine (T), Cytosine (C), Guanine (G), and Adenine (A).
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of these four bases: A, T, G and C. In the DNA double helix, the four chemical bases always bond with the same partner to form “base pairs” A always pairs with T; C always pairs with G.
In recent decades there have been significant advancements in the technology available for DNA sequencing. Broadly, there are two types of DNA sequencing:
shotgun and high-throughput. Shotgun (Sanger) sequencing is the more traditional approach, which is designed for sequencing entire chromosomes or long DNA strands with more than 1000 base pairs. It involves a rapidly expanding firing pattern to read the DNA in short fragments of 100 to 1000 base pairs, which are then overlapped with a computed analysis system.
High-throughput is the next-generation sequencing (NGS) method of DNA sequencing, which has led to the rapid acceleration of DNA sequencing and broadened knowledge in the field. It is able to produce thousands of sequences simultaneously, which lowers the cost of the technique significantly.
New methods are still under development, including some that utilize nanopores to sequence the DNA. This would work by threading single strands of DNA through nanopores in the cell membrane, which would then be read by the technology in single file.
DNA sequencing may be used to determine the sequence of individual genes, larger genetic regions, full chromosomes, or entire genomes of any organism. This information is useful for researchers in understanding the type of genetic information that is carried in the DNA, which may affect its function in the body. The human genome contains about 3 billion base pairs that spell out the instructions for making and maintaining a human being. Applications such as whole genome sequencing or WGS allows complete coverage of the entire genomic material. This is in contrast to targeted sequencing where only specific regions of interest are sequenced.
Sequencing is the procedure of determining the order of nucleotides in a DNA section comprising a gene. Next-generation sequencing is a technology for determining the sequence of DNA or RNA to study genetic variation associated with diseases or other biological phenomena.
This method was initially called “massively parallel sequencing”, because it enabled the sequencing of many DNA strands at the same time, instead of one at a time as with traditional Sanger sequencing by capillary electrophoresis.