Genomic Profiling of Hematologic Malignancies
Hematologic cancer account for a significant fraction of new cancer cases. The many stages of hematopoietic differentiation and abundant opportunities for mutations give rise to a vast phenotypic and genetic heterogeneity of acute and chronic myeloid malignancies. The ability to accurately characterize diseased samples will allow more differentiated disease classification, risk stratification, better understanding of disease mechansims, and improved therapeutic decisions.
The hematology community have relied mainly on cytogenetic and single-gene methods in the past. Next-generation sequencing (NGS) provides many benefits over traditional methods, including faster, more scalable, and more sensitive profiling of hematologic tumors. Amplicon-based targeted sequencing is an ideal approach that is cost-effective and easy to implement in the laboratory. The workflow is fast and simple to carry out, and the approach allows researchers to focus only on genes related to one or more subtypes of hematologic malignancy to control the cost of sequencing.
CleanPlex® NGS Panels – Versatile Solution for Genomic Profiling of Hematologic Tumors
Obtain Accurate, Reproducible, and Highest-Quality Results from Tumor Samples
CleanPlex Custom NGS Panels are powered by Paragon Genomics’ CleanPlex Technology – an ultra-high multiplex PCR-based target enrichment technology for next-generation sequencing (NGS). It features a highly advanced proprietary primer design algorithm and an innovative, patented background cleaning chemistry.
Researchers pick targets or genes that they would like to interrogate, and our experts design and deliver the custom assays in complete kit format (input to sequencing-ready NGS libraries) in just 4 to 6 weeks.

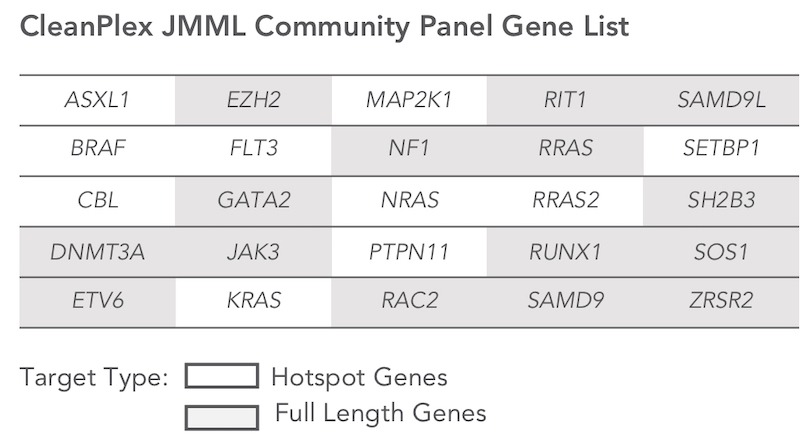
Prof. Elliot Stieglitz‘s Lab at the UCSF Benioff Children’s Hospital developed a 25-gene custom NGS assay powered by Paragon Genomics’ CleanPlex technology to retrospectively measure the molecular burden of juvenile myelomonocytic leukemia (JMML) patients at the time of initial diagnosis and posttherapy before hematopoietic stem cell transplantation. DNA was extracted from frozen cryopreserved mononu- clear cells or sections of paraffin-embedded bone marrow blocks for targeted library preparation. In their publication, the researchers demonstrated that molecular testing can be helpful to distinguish between responders and nonresponders and should become an integral part of clinical care.
When we decided to choose a sequencing platform, we had several choices and I am delighted that we chose Paragon Genomics. Paragon Genomics has enabled us to move our sequencing in-house with an affordable and easy-to-use product. They assisted us every step along the way from choosing the amplicons, to guiding us through library preparation to assisting with analysis. I strongly recommend their CleanPlex® amplicon-based targeted sequencing approach and could not be happier with their product.
Free Custom/Community Panel Consultation
High-Performance Genomic Profiling Solution for JMML and Other Hematologic Malignancies (AML, ALL, CML, NHL, MM, etc.)
CleanPlex Community NGS Panels are developed by Paragon Genomics or in collaboration with researchers around the world. To access Dr. Stieglitz’s CleanPlex JMML Community Panel or to create a similar custom panel to study hematological tumors, schedule a free consultation below by entering your email address.
The CleanPlex JMML Community Panel targets 25 genes that are frequently mutated in JMML.
At Paragon Genomics, we take your privacy and data very seriously. You can review our Privacy Policy online or download the PDF.