Targeted Gene Fusion Detection Technology for NGS
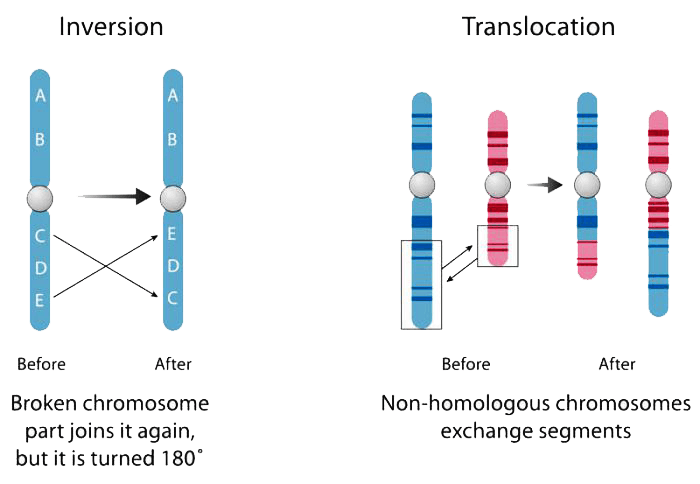
A gene fusion is a hybrid gene formed from two previously independent genes. It can occur as a result of translocation, interstitial deletion, or chromosomal inversion. Many gene fusion mutations are highly characterized and are a major cause of cancer, making accurate detection critical to research and to clinical applications. Fluorescence in situ hybridization (FISH) has been the gold standard for fusion detection, however it can only detect single mutations at a time and cannot detect novel fusion gene partners. RNA-seq is another method for fusion detection, but has low sensitivity and a high read requirement as a result of whole transcriptome sequencing.
At Paragon Genomics, we have created a targeted multiplex fusion detection technology that can also detect novel fusion gene partners. Built upon our CleanPlex® technology, our fusion technology allows for targeted interrogation of just the genes of interest in order to increase sensitivity, reduce non-specific reads, and allow accurate fusion detection in one simple workflow.
Known and Novel Fusion Detection (Gene Fusion)
Paragon Genomics’ RNA fusion technology utilizes the CleanPlex amplicon sequencing chemistry for confident detection of known and novel fusions using two routes. Both methods use a proprietary multiplex PCR background cleaning chemistry to effectively remove non-specific PCR products, resulting in clean and high-quality libraries (Figure 3) for best-in-class target enrichment performance and efficient use of sequencing capacity.
AccuFusionTM
- Dual primer multiplex PCR design
- Targeted known fusion detection
- Little amplification bias and high copy number correlation
- AccuFusion RNA Lung Cancer Panel includes fusion genes associated with ALK, CIT, EML4, FGFR1, MBIP, NRG1, NTRK1, NTRK3, PDGFRA, RET, ROS1, TACC3.
OmniFusionTM
- Single primer multiplex PCR design with template switching chemistry
- Ability to detect novel fusion gene partners
- Efficient sequencing with low rRNA rates
- OmniFusion RNA Lung Cancer Panel include fusion genes associated with 11 cancer driver genes ( ALK, CIT, MBIP, MET, NRG1, NTRK1, NTRK3, PDGFRA, RET, ROS1, TACC3).
Both methods are highly sensitive and compatible with low quality RNA such as FFPE, and other sample types such as fresh/frozen tissue or cell lines. The chemistry also allows for minimal input samples, and can generate robust libraries for confident calls from as little as 25ng (OmniFusion) or 10ng (AccuFusion). In addition to our expanding collection of ready-to-use panels, customized panels can be designed based on characterized breakpoints under the AccuFusion workflow, or based on specific genes of interest to discover new fusion events under the OmniFusion single primer workflow.
Fast, Easy, Single-Tube Workflow
The workflows for both methods are similar. From purified RNA samples to sequencing-ready libraries, samples can be completed in 6 hours (Figures 1A, 1B) with about 75 minutes of hands on time. The extracted RNA is first converted to cDNA by reverse transcription.
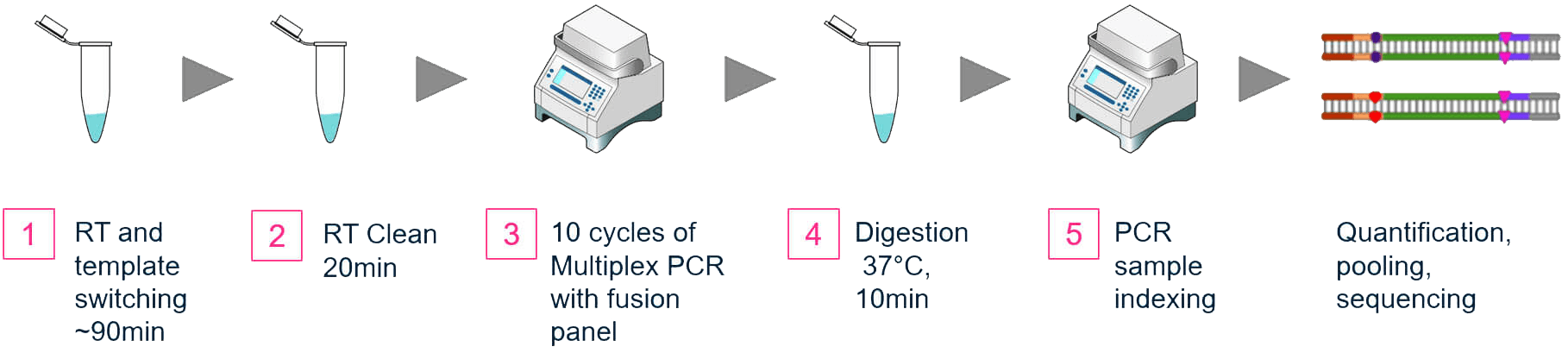
Figure 1A. OmniFusion Single-Primer Gene Fusion Detection Workflow
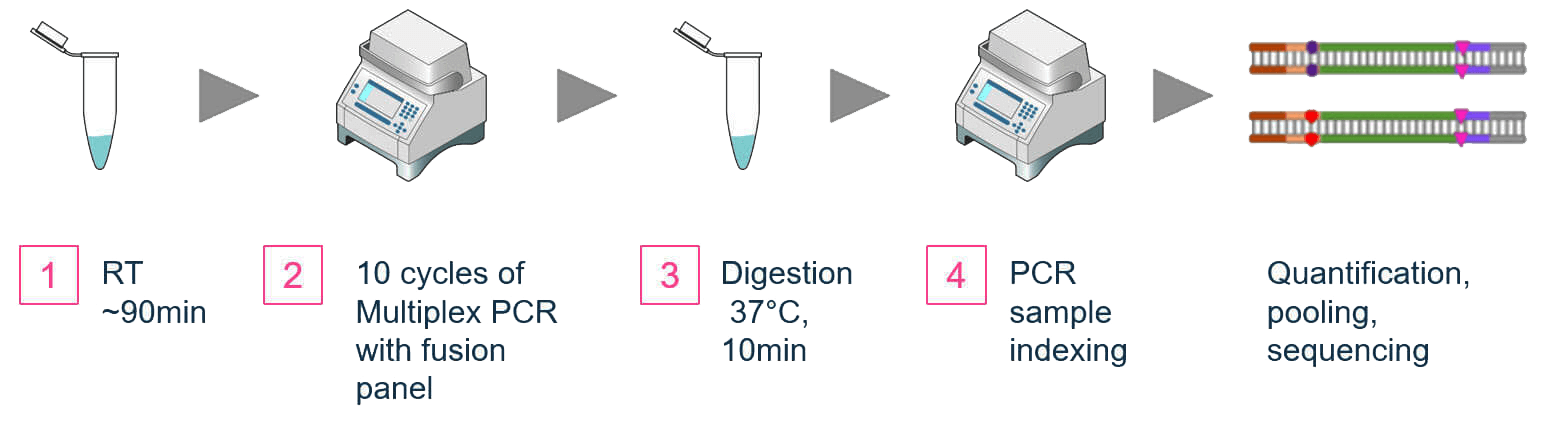
Figure 1B. AccuFusion Dual-Primer Gene Fusion Detection Workflow
Uniquely in the OmniFusion workflow, a universal primer is added to the cDNA by template switching. Multiplex PCR follows with target specific primers for the total amplification (Figure 2).
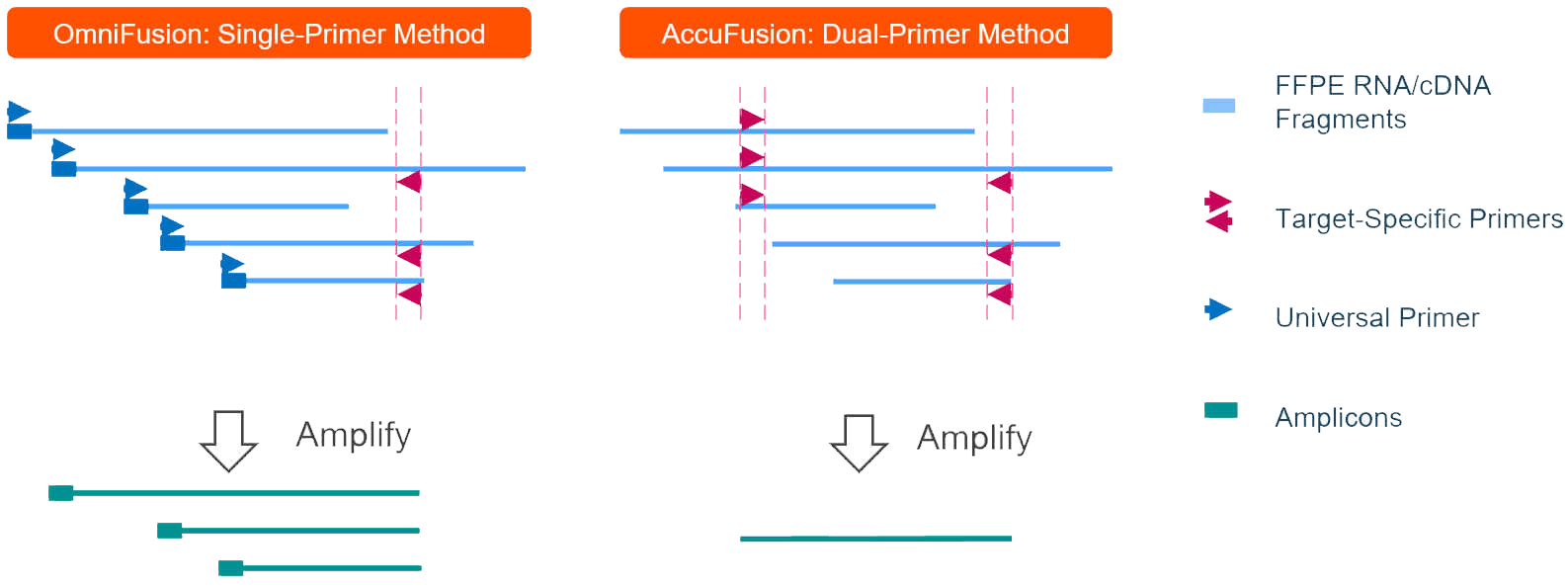
Figure 2. AccuFusion vs. OmniFusion Method Comparison
The last PCR step attaches sequencing specific indexes for sequencing on both Illumina and Ion Torrent platforms. Combined with our index offerings, 384 samples can be pooled for sequencing on a single flow cell with our unique-dual-index set for Illumina, or 2688 samples with our combinatorial dual indexes for Illumina.
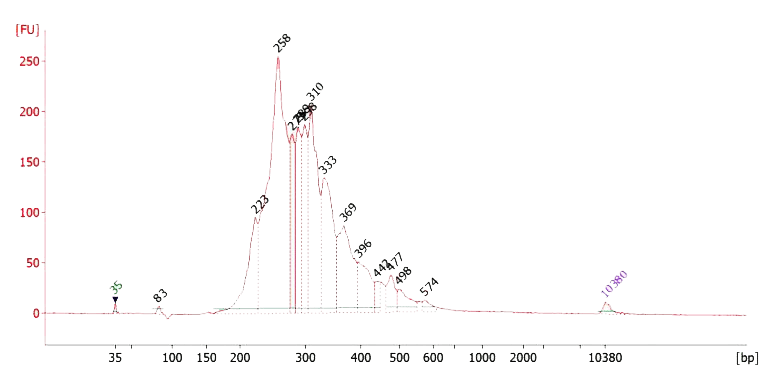
Figure 3. OmniFusion RNA Fusion Library Trace
Lung Cancer Panel Performance
Two Lung Cancer Fusion panels (AccuFusion™ RNA Lung Cancer Panel, OmniFusion™ RNA Lung Cancer Panel) with 61 unique fusion targets were created with the OmniFusion and AccuFusion methods. The panels were run with control RNA fusion samples to evaluate performance.
- Accuracy: All 10 fusions were detected in Seraseq® Fusion RNA Mix v4 using AccuFusion, whereas an additional novel fusion target was also detected with the OmniFusion workflow (Figure 4).
- Specificity for Cost Effective Sequencing: Combined with the power of CleanPlex chemistry and reverse transcription optimization, both gene fusion detection methods generate high on-target and mapping rates, and minimum ribosomal RNA (rRNA) and primer-dimer rates for efficient sequencing of only reads you want, and none that you don’t (Figure 5).
- Unbiased: The panels exhibit high correlation between observed fusion reads from sequencing and fusion copies as determined by dPCR, allowing additional confident quantitative analysis without the need for molecular barcoding and associated complex downstream analysis (Figure 4).
- Flexibility: Together with the single primer design combined with our proprietary ParagonDesignerTM algorithm, custom designs and panels can be generated in as short as 2 weeks for detecting novel and/or known fusion genes.
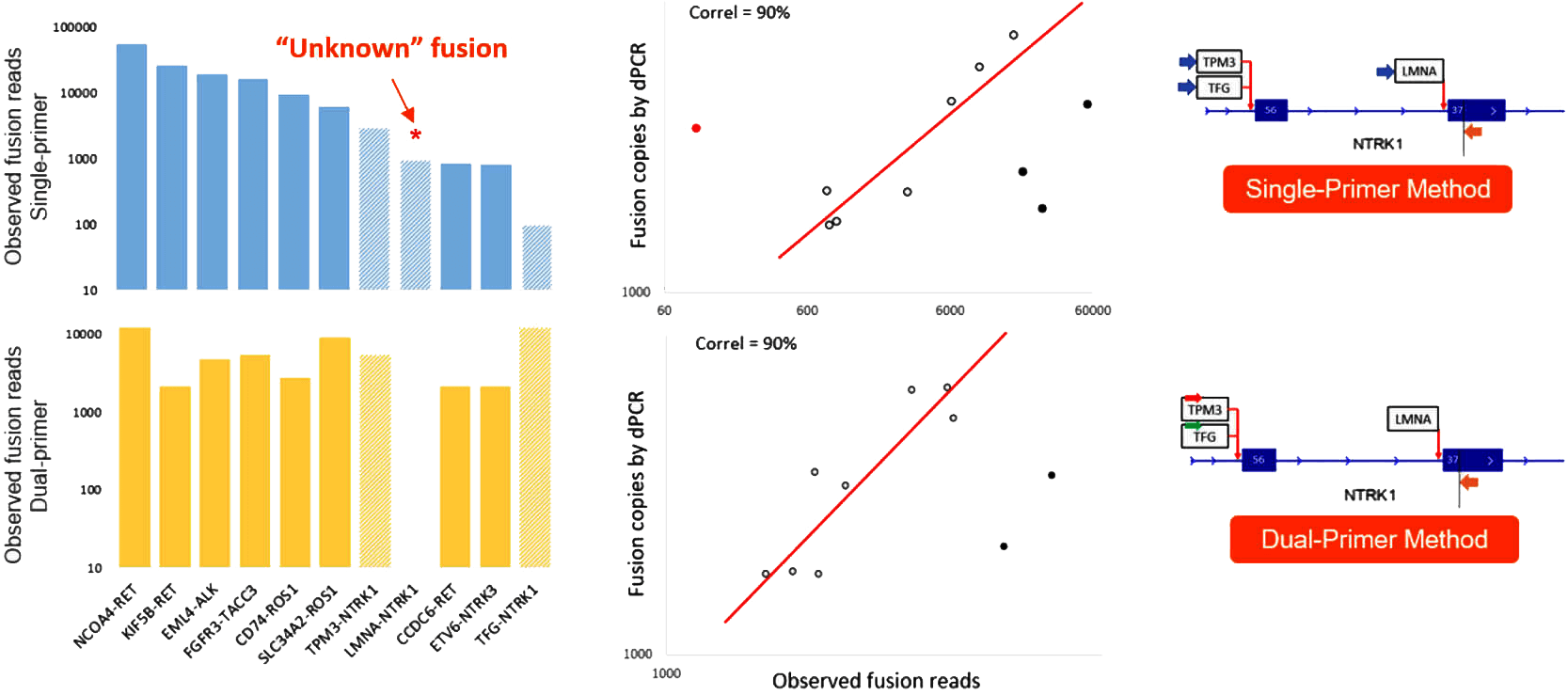
Figure 4. Seraseq® Fusion RNA Mix v4 (25ng) were used to generate concordance data with OmniFusion and AccuFusion Lung Cancer Panels. The generated libraries were sequenced at 0.1M reads per sample. The single-primer method can identify novel fusions with unknown information (i.e., LMNA-NTRK1 fusion). The dual-primer method can be used to interrogate the presence of known fusion targets (i.e., TPM3/TFG-NTRK1 fusions).
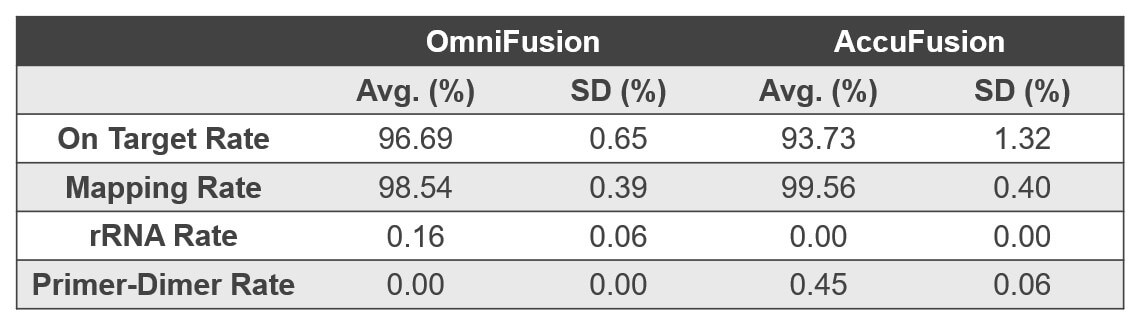
Figure 5. Highly specific amplification and minimum non-specific product in the final libraries for increased sample multiplexing and cost effective sequencing.
Order a Free Trial Kit to Test
View other data on CleanPlex® technology for amplicon sequencing
FAQ about Gene Fusion
A gene fusion is a hybrid gene formed from two previously independent genes. It can occur as a result of translocation, interstitial deletion, or chromosomal inversion. Gene fusions are clinically relevant to some types of solid tumors but impact mostly hematological cancers. The identification of these fusion genes plays a prominent role in being a diagnostic and prognostic marker.
In recent years, next generation sequencing technology has become available to screen known and novel gene fusion events on a genome wide scale. Until the advent of RNAseq (paired-end sequencing), chromosome banding analysis, fluorescence in situ hybridization (FISH), and reverse transcription polymerase chain reaction (RT-PCR) were the most commonly used methods to identify gene fusions. These methods all have their distinct shortcomings. The high-throughput nature of RNAseq and its relatively low cost combined with the much greater information available from the technique make it an attractive alternative to all other methods.
Gene fusion plays an important role in cancer. Fusion genes can contribute to tumor formation because fusion genes can produce much more active abnormal protein than non-fusion genes. Oncogenic fusion genes may lead to a gene product with a new or different function from the two fusion partners. Most fusion genes are found from hematological cancers, sarcomas, and prostate cancer. Oncogenic fusion transcripts may also be caused by trans-splicing or read-through events.
A gene is a region of DNA that encodes function. It is a basic physical and functional unit of heredity. A chromosome consists of a long strand of DNA containing many genes. A human chromosome can have up to 500 million base pairs of DNA with thousands of genes. In humans, genes vary in size from a few hundred DNA bases to more than 2 million bases. The Human Genome Project estimated that humans have between 20,000 and 25,000 genes. Every person has two copies of each gene, one inherited from each parent. Some genes act as instructions to make molecules called proteins. However, many genes do not code for proteins.
