What is Targeted Sequencing (Resequencing)?
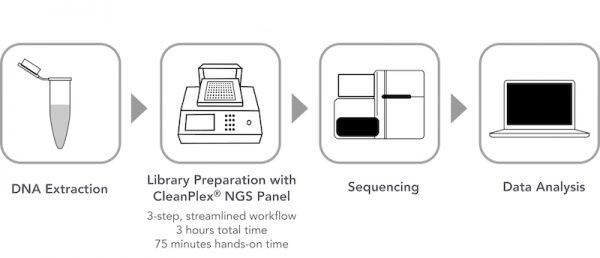
Targeted Sequencing or Resequencing is a method for only sequencing part of a whole genome or regions of interest without sequencing the entire genome of a sample. In order to only focus on specific or clinically relevant regions of a genome or DNA sample, it requires a pre-sequencing DNA preparation step called Target Enrichment where target DNA sequences are either directly amplified (amplicon or multiplex PCR-based) or captured (hybrid capture-based) and then subsequently sequenced using DNA sequencers. Below is a typical targeted sequencing workflow. (Figure 1)
Figure 1. A Typical Targeted DNA Sequencing Workflow
Why Targeted Sequencing or Targeted Gene Sequencing?
Whole genome sequencing (WGS) and its corresponding whole genome amplification (WGA) method are more suitable for research and discovery types of applications, while targeted sequencing or targeted gene sequencing is essential to many fast-growing clinical and industrial applications where cost and speed are more important.
One of the challenges the genomics community faces is the continued acquisition of large amounts of sequencing raw data that is yet to be fully and successfully translated and interpreted to help advance research, diagnose and cure diseases on a wider scale. Even with current reduced sequencing costs, a whole genome sequencing approach can be practically used only in specific scenarios such as basic research, population genetics or rare disease detection. A focused or targeted approach would be more appropriate for understanding disease progression and guiding therapy selection in clinical setting or massively screening DNA samples in industrial applications. In addition, sequencing an entire genome or exome can be prohibitively expensive in terms of laboratory operations and bioinformatics infrastructure for storing and processing large amounts of data. Therefore, targeted sequencing has become vital for the continued progress of precision medicine and research.
The following table compares WGS and targeted sequencing in terms of sequencing and library preparation reagent costs. In addition to reagent costs, bioinformatics analysis time and cost shall also be taken into account when considering the suitable methods for your applications.
| Comparison of WGS and Targeted Sequencing Methods | |||
| WGS | Targeted Sequencing | ||
| Human Whole Genome | Human Whole Exome (50Mb) | Targeted Panel (1Mb) | |
| Target Region Size (basepair) | 3 x 109 | 5 x 107 | 1 x 106 |
| Depth of Coverage | 30X | 100X | 1000X |
| Number of Samples per Sequencing Run* | 8 | 145 | 725 |
| Cost of Sequencing Reagent per Sample** | $1,500 | $100 | $15 |
| Cost of Target Enrichment per Sample*** | N/A | $250 | $100 |
| Total Cost per Sample (excluding bioinformatics) | $1,500 | $350 | $115 |
* Based on Illumina HiSeq 2500 System dual flow cell high output mode
** Based on Illumina HiSeq SBS V4 cost
*** Based on average target enrichment and library preparation kit prices
The clear benefits of targeted resequencing are driving the adoption of NGS in areas such as translational research, clinical diagnostics and industrial applications while the value also shifts from sequencing reagents to target enrichment reagents on a per sample basis. For a small targeted sequencing panel covering for example only 100 kb region of interest, most of the cost is incurred at the library preparation and target enrichment steps while the cost of sequencing reagents is negligible for each given sample if deep sequencing is not required.
What are major applications of Targeted (Gene) Sequencing?
Currently, targeted sequencing has been applied to many areas from basic research to clinical diagnostics and applied markets. Below is a list of major applications.
- Cancer
- Cancer research and diagnostics
- Tumor profiling
- Cancer liquid biopsy
- Immuno-Oncology
- Minimal residual disease testing
- Circulating Tumor Cell (CTC) analysis
- Reproductive Health
- Carrier screening
- Non-invasive prenatal testing
- Preimplantation genetic diagnosis (PGD)
- Newborn screening
- Cardiovascular Disease Testing
- Infectious Disease Testing and Surveillance
- Transplant Genomics / HLA typing
- Inherited Disease Testing
- Metabolic and Immune Disorders
- Neurological Disease Testing
- Companion Diagnostics
- Applied / Industrial Applications
- Agrigenomics / molecular breeding
- Food safety and animal health
- Forensics
- Environmental research
Introducing CleanPlex – the most advanced amplicon-based targeted sequencing technology
CleanPlex® is an ultra-scalable and ultra-sensitive amplicon-based targeted sequencing technology. It features a highly advanced proprietary multiplex PCR primer design algorithm, an exceptionally uniform multiplex PCR amplification chemistry and an innovative, patented background cleaning chemistry. Together, they allow CleanPlex Ready-to-Use and Custom NGS Panels to break the limits of traditional amplicon-based and hybrid capture-based targeted sequencing technologies.
Feature Highlights:
- Super high amplification uniformity and super low PCR background noise (more accurate variant calling or less sequencing cost)
- Single-tube and 3-hour workflow with minimal hands-on time (easy automation)
- Compatible with difficult samples (degraded FFPE DNA, FFPE RNA, cfDNA, cfRNA) and major sequencing platforms (Illumina, Ion Torrent, Genapsys, MGI DNBSeq)
- Extreme sensitivity (down to single cell level direct amplification*)
- Excellent panel size scalability from a few to over 20,000 amplicons in a single multiplex PCR pool
- Detection of single nucleotide variants (SNVs), small insertions and deletions (Indels), copy number variations (CNVs), gene fusions / splice variants, expression level, tumor mutational burden (TMB), microsatellite instability (MSI), internal tandem duplication (ITD), etc.
*based on data from our collaborators RareCyte and Mayo Clinic
Below is a typical workflow (Figure 2) of CleanPlex DNA target enrichment and library preparation. The CleanPlex DNA workflow involves 3 simple steps, each consisting of a thermal cycling or incubation reaction followed by a library purification using magnetic beads. The streamlined protocol can be completed in just 3 hours. In step 1, targets of interest are amplified in a multiplex PCR reaction. In step 2. primer-dimers, non-specific PCR products, and complex molecular-debris are biochemically removed in a digestion reaction featuring the innovative and proprietary CleanPlex Background Cleaning chemistry. In step 3, libraries are barcoded with sample indexes via a PCR indexing reaction. The input materials could be genomic DNA and DNA extracted from FFPE, fresh/frozen tissues or blood liquid biopsies. By adding a reverse transcription step in the front, the input material could be RNA too. (Figure 3)
Figure 2. CleanPlex DNA Target Enrichment and Library Preparation Workflow
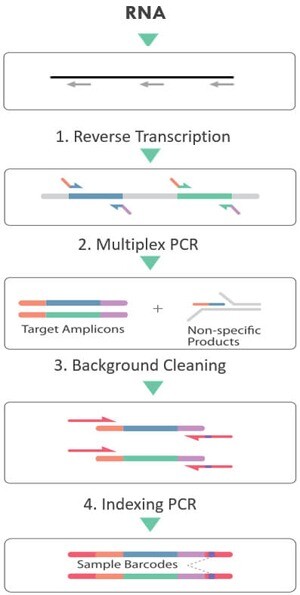
Figure 3. CleanPlex RNA Target Enrichment and Library Preparation Workflow
Discover more with less™
The combination of superior primer design and innovative multiplex PCR-based targeted sequencing and library preparation chemistry give rise to CleanPlex NGS Amplicon Panels’ ultra-high multiplexing capability, high performance, low input requirement, high sensitivity, single-tube workflow, and cost-effective amplicon sequencing. These remarkable features and benefits allow researchers and assay developers to discover more with less.
Discover More
Discover more with CleanPlex NGS Panels
• Multiplex 20,000+ amplicons per reaction
• High target design rate
• High coverage uniformity
• High on-target rate
• High sensitivity (1% LOD with 10 ng input)
Use Less
Use less input and resources to reduce costs
• Inputs as low as 6 pg (single cell gDNA)
• Fast 3-hour protocol
• Simple, streamlined workflow
• Efficient use of NGS reads
Request a Free Targeted Sequencing Consultation
Have specific requests or questions? Schedule a free consultation with our PhD-level expert scientists to learn how our best-in-class targeted sequencing custom panel design services can help you advance your work.
View more data on CleanPlex® technology for targeted sequencing
FAQ About Targeted Sequencing or Targeted Gene Sequencing
Targeted sequencing or targeted gene sequencing is a fast and low-cost way to detect known and novel variants in selected (targeted) sets of genes or genomic regions.
DNA sequencing can be accomplished using several different methods. By leveraging current genomic knowledge, a targeted NGS approach utilizes molecular biology methods to enrich for specific genetic sequences, to focus studies on individual genes or genomic regions. This targeted DNA sequencing approach improves coverage, simplifies analysis and interpretation, lowers total sequencing workflow costs. This targeted approach allows to sequence at a much higher depth of coverage to discover rare variants.
There are two methods of targeted sequencing to enrich for specific genetic regions: Hybridization capture and Amplicon-based enrichment.
Hybridization capture can be either solution-based or performed on a solid-substrate such as a microarray. Both methods require the use of synthesized oligonucleotide probes (also known as baits) that are complementary to the genetic sequence of interest. In solution-based methods, the probes are biotinylated and added to the genetic material in solution to hybridize with the desired regions of interest. Magnetic streptavidin beads are used to capture and isolate the hybridized probes from the unwanted genetic material. With array-based capture, the probes are attached directly to the solid surface. The genetic material is applied to the microarray and target regions hybridize to the surface. Any unbound material is washed away, leaving the desired target regions isolated on the substrate. For both methods, the isolated and purified target regions are subsequently amplified and prepared for sequencing.
Amplicon-based enrichment uses carefully designed PCR amplicons to flank targets and specifically amplify regions of interest. The amplified products are then purified from the sample material and used for sequencing, bypassing the need for enrichment by hybridization.
Targeted gene sequencing panels are useful tools for analyzing specific mutations in a given sample. Focused panels contain a select set of genes or gene regions that have known or suspected associations with the disease or phenotype under study. Gene panels can be custom designed to include genomic regions of interest.
Target enrichment is a cost-effective and efficient method to capture specific genomic regions of interest after library preparation for NGS. Target enrichment enables focused sequencing resources, which leads to simplified analysis. The technique increases the probability of sequencing DNA fragments of the targeted regions compared to all other genes in the genome. As a result, the rest of the genome can be disregarded, simplifying downstream bioinformatics analysis. It also provides greater sequencing depth, which helps prevent false interpretations of sequencing data. It has many advantages over whole genome sequencing.
Targeted NGS is a type of sequencing that focuses on specific areas of the genome. Popular methods of targeted NGS include hybridization capture and amplicon sequencing. Targeted next generation sequencing allows you to sequence specific areas of the genome for in-depth analyses more rapidly and cost-effectively than whole genome sequencing. It offers the scalability, speed, and resolution to evaluate targeted genes of interest. Multiple genes can be assessed across many samples in parallel, saving time and reducing costs associated with running multiple separate assays. Targeted gene sequencing also produces a smaller, more manageable data set compared to broader approaches such as whole-genome sequencing, making analysis easier.