Amplicon-based Molecular Barcoding for Detecting Low-Frequency Alleles
CleanPlex® UMI Technology is a multiplex PCR or amplicon-based NGS targeted sequencing technology for precise tumor DNA analysis. It features a highly advanced proprietary primer design algorithm and an innovative, patented molecular barcoding chemistry. Together, they allow CleanPlex UMI Ready-to-Use and Custom NGS Panels to confidently detect low-frequency variants (circulating tumor DNA – ctDNA) in cell-free DNA (cfDNA).
CleanPlex UMI Target Enrichment Workflow
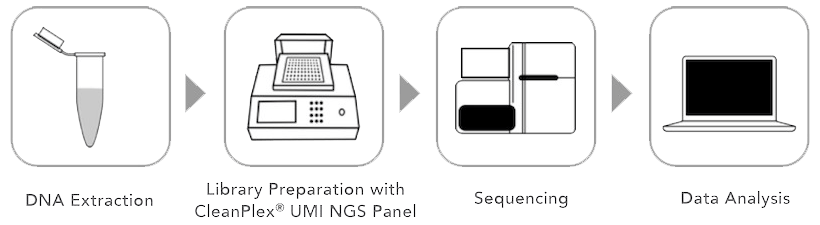
CleanPlex UMI multiplex PCR-based target enrichment workflow. CleanPlex UMI Technology allow molecular-barcoded and target-enriched NGS libraries to be easily and quickly prepared for sequencing.
Ultrafast and simple workflow
CleanPlex UMI Technology features a simple workflow that can be completed in 3.5 hours and requires only 85 min of hands-on time. Molecular-barcoded and target-enriched NGS libraries can be quickly prepared for faster time to results.
CleanPlex UMI Technology with Unique Molecular Identifier
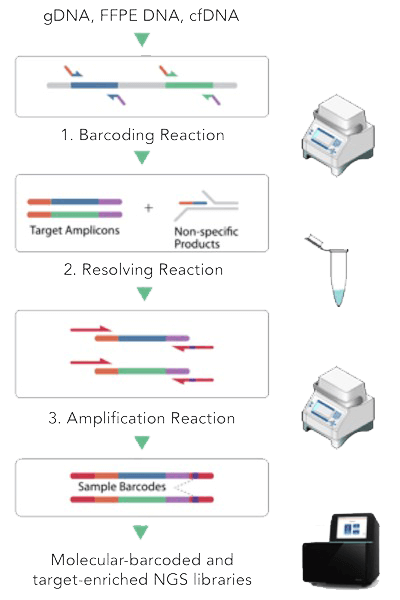
CleanPlex UMI targeted library preparation protocol. The CleanPlex UMI protocol involves 3 simple steps, each consisting of a thermal cycling or incubation reaction followed by a library purification using magnetic beads. The streamlined protocol can be completed in just 3.5 hours. The only instrument required to complete the protocol is a thermal cycler.
Innovative molecular barcoding chemistry
CleanPlex UMI Technology uses a proprietary multiplex PCR-based molecular barcoding chemistry to uniquely label each DNA molecule. The incorporated unique molecular identifiers (UMIs) allow PCR and sequencing errors to be corrected in the sequencing data, resulting in improved sensitivity and specificity for variant detection.
CleanPlex UMI (Molecular Barcoding) Chemistry
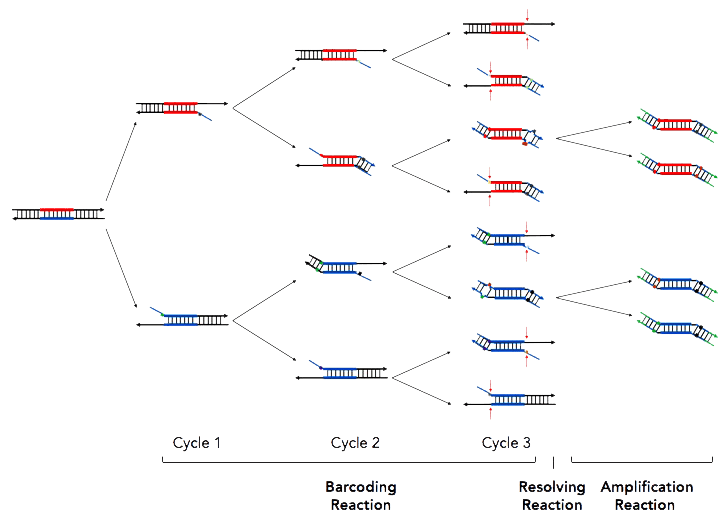
CleanPlex UMI molecular barcoding chemistry. The first step of the CleanPlex UMI workflow is a multiplex PCR reaction that uses UMI-labeled target-specific primers to barcode and amplify targets of interest. The second step is a biochemical reaction that resolves the correct UMIs by removing PCR products carrying redundant and partial UMIs. The last step is an amplification reaction that uses CleanPlex Unique Dual-Indexed PCR Primers, which ensures sample demultiplexing with high accuracy, to amplify and add sample-level indexes to the NGS libraries. CleanMag® Magnetic Beads are recommended for library purification.
Confident variant detection with UMI error correction
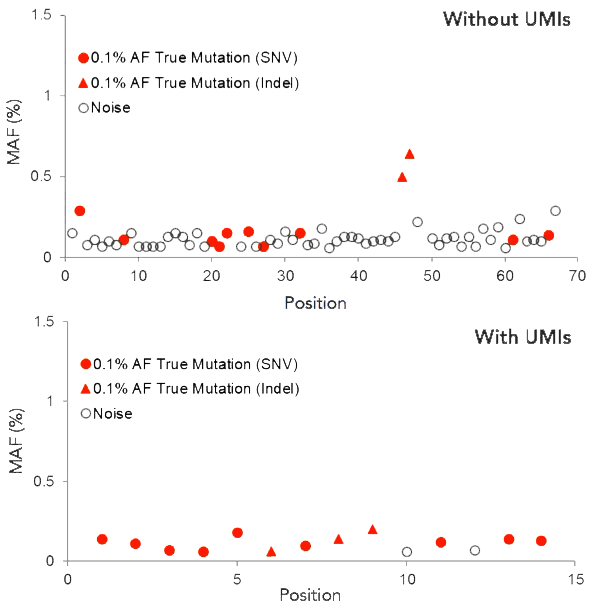
Accurate variant calling enabled by UMIs. 50 ng of SeraCare Seraseq™ ctDNA Complete™ Mutation Mix at minor allele frequency (MAF) of 0.1% was used to prepare targeted NGS libraries using the CleanPlex UMI Lung Cancer Panel, which covers 13 mutations present in the reference material. The resulting data was analyzed with and without the use of UMIs. The bottom panel demonstrates that UMI-enabled error correction results in a significant reduction in false positives, allowing true mutations (orange circles and triangles) to be distinguished from background noise (white circles).