High quality libraries from low-input and challenging samples
The streamlined CleanPlex® workflow minimizes sample loss to preserve genomic information in low-input and challenging samples, such as degraded FFPE DNA from tumor samples, cell-free DNA from liquid biopsies and single circulating tumor cell (CTC) from blood. High-quality target-enriched libraries can be generated from as little as 1 ng of DNA and even down to 6 pg of DNA from a single CTC.
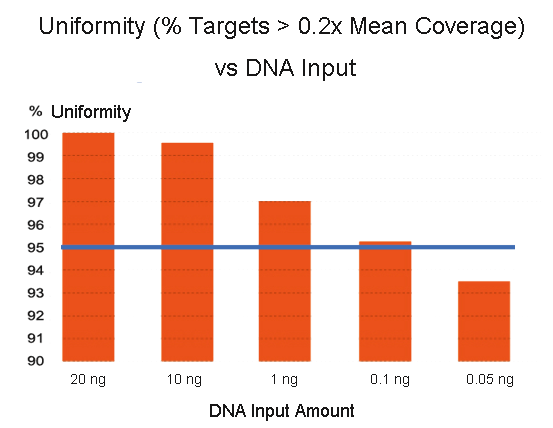
High performance even with low input amounts. Libraries were prepared with the CleanPlex OncoZoom® Cancer Hotspot Panel (601 amplicons) using varying amount of input DNA and sequenced on an Illumina platform. CleanPlex technology was able to produce libraries with >95% coverage uniformity (measured as % covered by at least 0.2X mean coverage) using as little as 50 pg of input genomic DNA.
Single cell targeted DNA sequencing with CleanPlex OncoZoom Cancer Hotspot Panel
CleanPlex NGS Panels can directly amplify single cells without pre-amplification such as whole genome amplification (WGA) while still maintaining high amplification uniformity. In addition, this avoids WGA bias and has much lower false positives and false negatives in terms of variant calls.
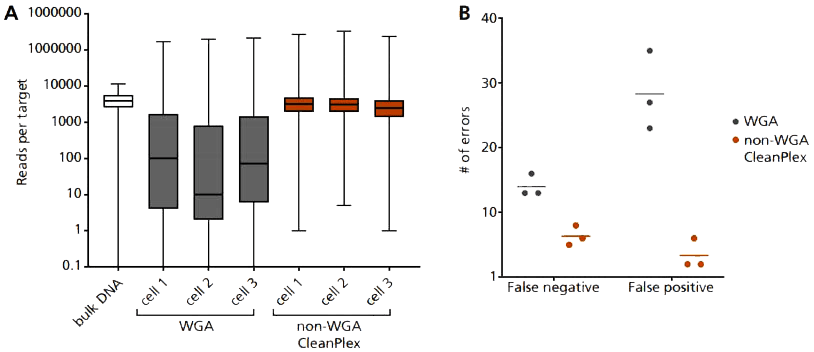
Single-cell targeted DNA sequencing with CleanPlex OncoZoom. Single-cell lysate obtained from RareCyte’s RareCyte’s CTC assays and CyteFinder Instrument is input as template into Paragon’s CleanPlex OncoZoom Cancer Hotspot Panel, with modified primer concentration, PCR cycle number, and clean-up steps to compensate for low-input DNA concentration. Using this non-WGA method vastly improves: (A) coverage uniformity, and incidence of (B) false negative and false positive errors, when compared to single- cell WGA products.
Detect variants with high confidence (without unique molecular identifiers – UMIs)
CleanPlex NGS Panels are powered by high-fidelity PCR enzymes and generate high-quality data with low error rates to allow confident and sensitive variant calling. CleanPlex NGS Panels can accurately detect mutations at 1% allele frequency using just 10 ng of input DNA.
High-quality sequencing data with low error rate. A library was prepared with the CleanPlex OncoZoom Cancer Hotspot Kit (610 amplicons) using 10 ng of Horizon Discovery HD780 cfDNA reference standard and sequenced on an Illumina platform to an average read depth of 8,500. A histogram of the frequency of random errors from sequencing shows that CleanPlex Kits generated high-quality data with the majority of random errors present at less than 0.2% frequency. This low level of background errors allows confident variant calling of mutations at 1% allele frequency.
High variant call concordance. Libraries were prepared with the CleanPlex OncoZoom Cancer Hotspot Panel (610 amplicons) using 10 ng of Horizon Discovery HD780 cfDNA reference standards and sequenced on an Illumina platform to an average depths ranging from around 1,100 to 9,800 reads per amplicon. The data shows that CleanPlex consistently detected validated variants at the expected frequency. The red lines represent the known allele frequencies of the eight mutations, and numbers are the averages of the detected frequencies for each allele with standard deviation error bars.