COVID-19 Coronavirus (SARS-CoV-2) Whole Genome Sequencing Kits for Viral Detection, Tracking and Surveillance
Check out the Latest SARS Product: Emerging Variants Add-on Panel v2 Covers all Characteristic Mutations of Omicron and its Sub-Lineages: BA.1, BA.2, BA2.12.1, BA. 2.75, BA.3, BA.4, and BA.5. Newest variants (XBB) can be identified via FLEX Kit.
Why COVID-19 / SARS-CoV-2 Whole Genome Sequencing (COVID-19 Sequencing or SARS-CoV-2 Sequencing)?
Whole genome sequencing is a critical tool in understanding emerging viruses. Next-Generation Sequencing (NGS) can track COVID-19 transmission and viral mutations, inform testing protocols and infection control measures, and guide vaccine and therapeutics development.
COVID-19 NGS (SARS-CoV-2 NGS) Method Comparison
| Amplicon Sequencing NGS (CleanPlex SARS-CoV-2 Panel or Flex Panel) | Hybrid Capture-based NGS | Metagenomics NGS | |
| Time of Library Prep and Target Enrichment | 5 hours | 1 Day | 4-6 hours |
| Sequencing Reads per Sample | ~ 100K to 500K | >5M | >10M |
| Max Genome Coverage at 10-20 viral copies* | > 98% | ~5-10% | Low |
| Mutation Tolerance | CleanPlex Flex Panel with degenerative primers captures new mutations | Fair | Good |
| Assay Sensitivity | High | Low | Low |
*Based on competitor published product sheets
Amplicon-based SARS-CoV-2 NGS Panels
- CleanPlex SARS-CoV-2 FLEX Kit (updated for detecting recent new mutations including N501Y mutation and 69/70 deletion)
- CleanPlex SARS-CoV-2 Kit
- SARS-CoV-2 Emerging Variants Panel Add-on v2
- CleanPlex Respiratory Virus Research Kit
- CleanPlex ACE2 & TMPRSS2 Kit
In March 2020, Paragon Genomics team quickly developed and launched an amplicon-based NGS panel: CleanPlex SARS-CoV-2 Panel for sequencing the whole genome of SARS-CoV-2 (the virus responsible for COVID-19) on both Illumina and MGI Tech Sequencing platforms. The panels are designed to obtain complete viral genomes even from samples with very low SARS-CoV-2 viral content.
Panel design is based on the SARS-CoV-2 sequence NC_045512.2. A total of 343 primer pairs, distributed in two pools, were selected by a proprietary panel design pipeline to cover the whole genome except for 92 bases at the ends. Primers were optimized to preferentially amplify the SARS-CoV-2 cDNA versus the background human cDNA or DNA. They were also optimized to uniformly amplify the covered genome. This expertly designed panel allows for the interrogation of the entire viral sequence with as little as 200,000 sequencing reads per sample.
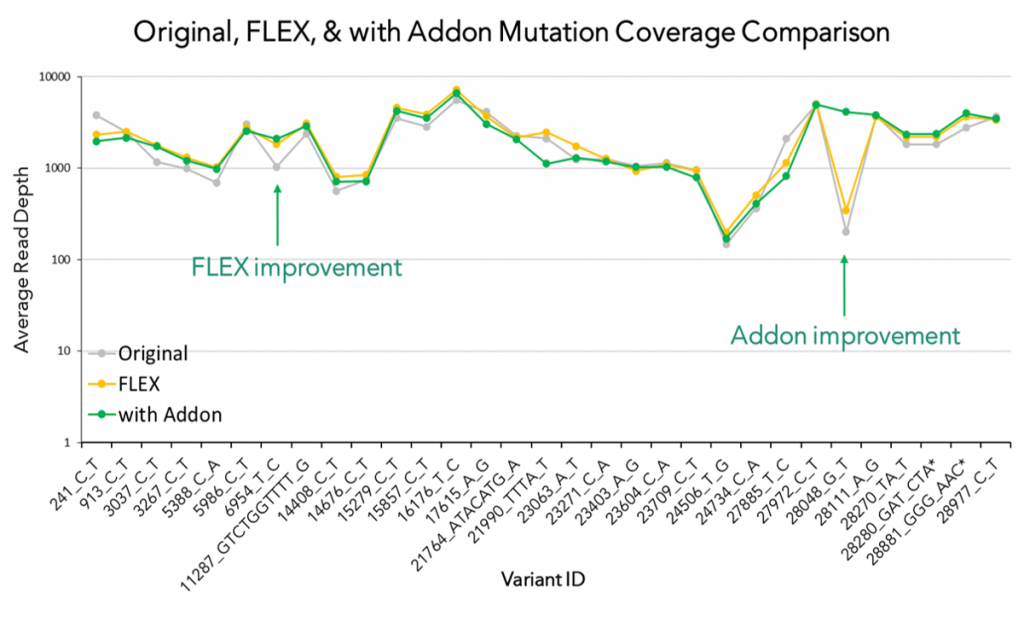
In addition, we also launched CleanPlex SARS-CoV-2 FLEX Kit and the Emerging Variants Panel Add-on for more robust coverage of the viral genome even as the virus mutates. The SARS-COV-2 FLEX panel differs from the standard SARS-COV-2 Panel in that it contains degenerate primer designs of polymorphic regions of the genome to allow more uniform amplification of variable strains. Recently, several SARS-CoV-2 mutations on the spike protein have been reported around the world, such that those found in the Alpha variant from United Kingdom (20B/501Y.V1, VOC 202012/01, or B.1.1.7 lineage), the Beta variant from South Africa (20C/501Y.V2, or B.1.351 lineage), the Gamma variant from Brazil (P.1 and P.2 ) and the Epsilon variant from West Cost United States (B.1.427 and B.1.429). Recently, one Brazilian lab using our SARS-CoV-2 panel was able to detect the newest variant of concern, variant B.1.617.2, also known as the Delta variant (Read Paper).
Emerging Variants Panel Add-on v2 (EVAv2) was launched in February 2022 as an update to the initial Add-on Panel, based on a systematic monitoring of the public health crisis and new variants of importance and concern. The omicron variant (B.1.1.529) was first discovered in November 2021 and contains several mutations in the receptor-binding domain (RBD) of the spike protein, which have been shown to significantly increase transmission rates and can negatively impact the efficacy of the recently developed vaccines and current COVID-19 qPCR mass testing methods. The Paragon team developed EVAv2 as a response to the emerging variant and optimized EVAv1 to maintain even coverage and confident identification of the defining mutations of the beta, delta, mu, and omicron variants: the panel has been confirmed in-silico to cover all defining and characteristic mutations of the sub-lineages of omicron: BA.1, BA.2, BA.3, BA.4, and BA.5.
Viral mutations can impact:
- Vaccine efficacy
- Transmission rates
- Disease severity
- Accurate viral detection
- Increased false-negative rates for COVID-19 testing
To date, the FDA has identified three molecular assays whose performance could be impacted by SARS-CoV-2 genetic variants and is suggesting that laboratories with access to quick-turnaround whole-genome sequencing services should consider further characterizing specimens with genetic sequencing. The CleanPlex SARS-CoV-2 FLEX Kit is a full genome sequencing-based COVID-19 assay for detection, research, and surveillance, with unique design elements that allow for more robust and confident variant calling, even when the virus mutates over time. Along with the Emerging Variants Panel Add-on v2, the panel was designed to confidently capture these new emerging mutations including critical N501Y mutation and 69/70 deletion along with other mutations specific to these new variants. You can download the Product Sheets here and here and read more about its application in our publications.
To differentiate SARS-CoV-2 virus from influenza A/B viruses in the flu season, we further launched CleanPlex Respiratory Virus Research Kit for respiratory virus research especially during flu seasons.
Research has shown that SARS-CoV-2 virus is internalized by the binding of its S protein to the host ACE2 receptor with the help of TMPRSS2 cell surface protein. The binding affinity of ACE2, along with ACE2 and TMPRSS2 expression levels, are major determinants of SARS-CoV-2 replication rate and disease severity. Due to this, we launched CleanPlex ACE2 & TMPRSS2 Kit to facilitate further investigations in host genome mutations related to the ACE2 binding affinity and expression of ACE2 and TMPRSS2.
“I was very impressed by how well the CleanPlex SARS-CoV-2 panel worked when we evaluated it back in March. Working with Paragon Genomics, we were able to leverage the panel to quickly launch a SARS-CoV-2 whole-genome sequencing assay and used it effectively for a number of genomic epidemiology studies. As a human geneticist and informaticist, I am also excited about the availability of the CleanPlex ACE2/TMPRSS2 germline + eQTL panel.”
“We have used the CleanPlex SARS-CoV-2 Panel for our COVID-19 wastewater surveillance program in Southern Nevada. The NGS panel has provided high sequencing coverage for emerging variants in our community and we are pleased with the ease and consistency of the workflow and results, respectively”
Submitted or Published Papers (>30+) Using CleanPlex® SARS-CoV-2 or FLEX Panel
- First Identification of the new SARS-CoV-2 Omicron Variant (B.1.1.529) in Italy (Read More)
- External quality assessment of SARS-CoV-2 Sequencing: an ESGMD-SSM Pilot Trial across 15 European Laboratories (Read More)
- SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools (Read More)
- First reported cases of SARS-CoV-2 sub-lineage B.1.617.2 in Brazil: an outbreak in a ship and alert for spread (Read More)
- Persistent SARS-CoV-2 infection and increasing viral variants in children and young adults with impaired humoral immunity (Read More)
- Genomic epidemiology of SARS-CoV-2 reveals multiple lineages and early spread of SARS-CoV-2 infections in Lombardy, Italy (Read More)
- Early pandemic molecular diversity of SARS-CoV-2 in children (Read More)
- Within-host diversity of SARS-CoV-2 in COVID-19 patients with variable disease severities (Read More)
- Characterization of local SARS-CoV-2 isolates and pathogenicity in IFNAR−/- mice (Read More)
- Pediatric COVID-19 in Southern California: clinical features and viral genetic diversity (Read More)
- The origin of SARS-CoV-2 in Istanbul: Sequencing findings from the epicenter of the pandemic in Turkey (Read More)
- Highly sensitive and full-genome interrogation of SARS-CoV-2 using multiplexed PCR enrichment followed by next-generation sequencing (Read More)
- Horizon Scanning COVID-19 Supplement High Impact Report Volume 1, Issue 1 (Read More)
- High Prevalence of SARS-CoV-2 Genetic Variation and D614G Mutation in Pediatric Patients with COVID-19 (Read More)
- Guidelines for Accurate Genotyping of SARS-CoV-2 Using Amplicon-Based Sequencing of Clinical Samples (Read More)
- Whole-genome Sequencing of SARS-CoV-2: Using Phylogeny & Structural Modeling to Contextualize Local Viral Evolution (Read More)
CleanPlex SARS-CoV-2 Amplicon Targeted Library Prep Workflow
The CleanPlex Coronavirus Panel workflow includes 3 simple steps that convert viral RNA to sequencing-ready libraries in 5 hours. CleanPlex® SARS-CoV-2 Panel is powered by Paragon Genomics’ CleanPlex Technology, which uses a proprietary multiplex PCR background cleaning chemistry to effectively remove non-specific PCR products, resulting in best-in-class target enrichment performance and efficient use of sequencing reads
Step 1 starts with isolated viral RNA input for reverse transcription. Paragon Genomics’ RT reagent utilizes a special mixture of optimized random primers to select RNA of interest and minimize transcription of other non-targeted materials such as rRNA from human cells.
Step 2 consists of the powerful multiplex PCR technology to simultaneously, uniformly, and efficiently amplify all regions of interest from the SARS-CoV-2 genome.
Step 3 is an indexing PCR that allows the addition of index primers for sample pooling and sequencing on either MGI or Illumina sequencing platforms.
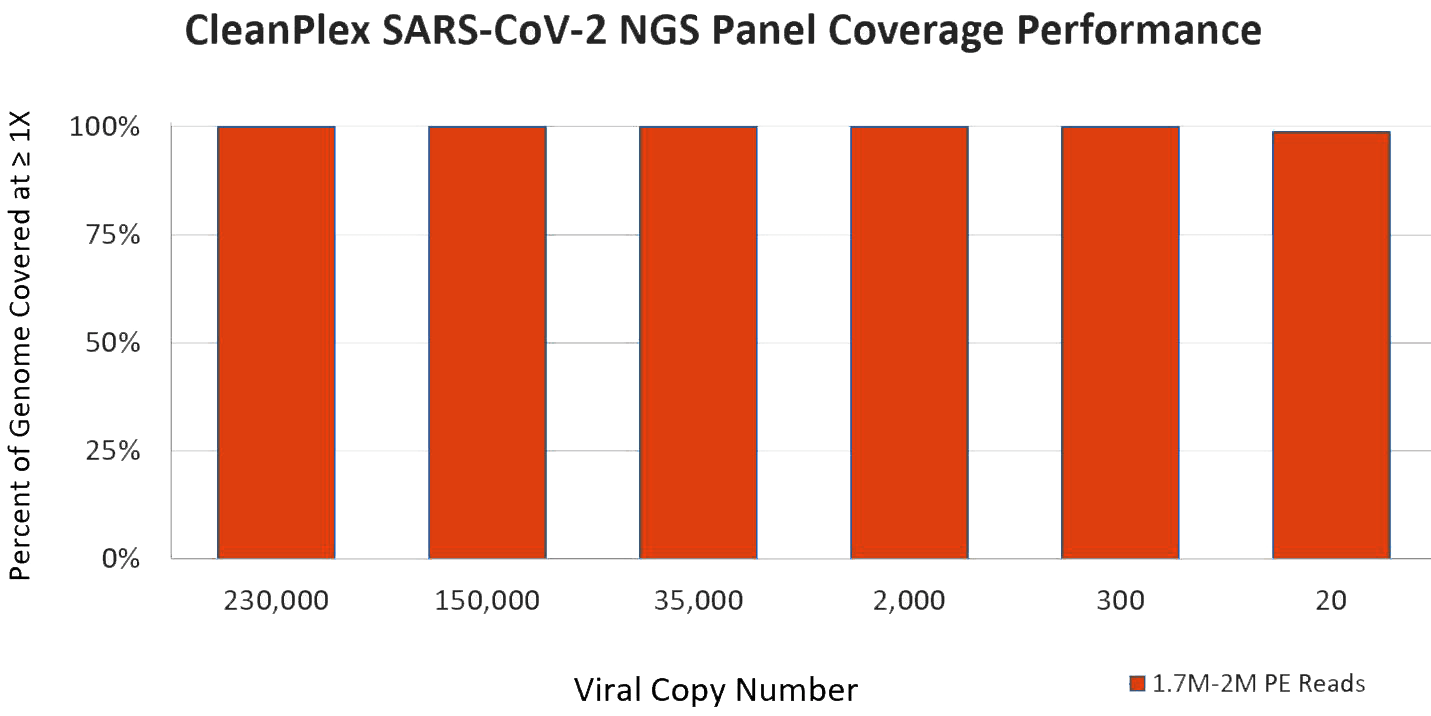
CleanPlex SARS-CoV-2 Panel Performance
Paragon Genomics’ CleanPlex SARS-CoV-2 NGS Panel shows exceptional sensitivity on very low virus copy numbers even at a low sequencing depth. Data below are shared by hospital collaborators using CleanPlex Panel to sequence patient samples that were tested positive by RT-PCR. With a high-throughput NGS sequencer from Illumina or MGI, you can pool hundreds to thousands of samples (only 0.2M reads needed for each sample) onto a single sequencing run.
To ensure that our customers get the most out of their sequencing runs, we’re proud to offer up to 2688 sample multiplexing capability with our combinatorial dual-indexed primers for Illumina sequencing. Additionally, we provide plated 384 unique-dual indexes, allowing for low variant calling and other in-depth sequencing applications. These exciting additions allow for cost-effective and high-throughput sequencing with our best-in-class CleanPlex targeted sequencing technologies.
Resources
Customer Testimonials on CleanPlex Infectious Disease Panels

Dr. Xiaowu Gai, Director of Bioinformatics at the Center for Personalized Medicine in the Department of Pathology and Laboratory Medicine at Children’s Hospital Los Angeles, said, “I was very impressed by how well the CleanPlex SARS-CoV-2 panel worked when we evaluated it back in March. Working with Paragon Genomics, we were able to leverage the panel to quickly launch a SARS-CoV-2 whole-genome sequencing assay and used it effectively for a number of genomic epidemiology studies. “

Scientists from Broad Institute’s Sabeti Lab worked with Paragon Genomics team to design a panel to sequence RNA viruses such as Ebola using CleanPlex and CleanPlex UMI technologies. The custom NGS assay features a single-tube workflow that is easy to perform and minimizes the risk for handling errors.
“We’re using Paragon’s CleanPlex and CleanPlex UMI technologies to sequence RNA viruses. These technologies enable us to rapidly and cost-effectively obtain complete viral genomes from clinical samples with low viral content. Further analyses of viral genomes, including the identification of minor variants, elucidates viral diversity and evolution. Paragon has enthusiastically worked with us to tackle this new application; they have diligently designed panels to our specifications, meeting with us frequently to optimize the design.” – Scientists from Broad Institute’s Sabeti Lab
Partnership
SOPHIA GENETICS AND PARAGON GENOMICS JOIN FORCES AGAINST THE COVID-19 CORONAVIRUS PANDEMIC
At Paragon Genomics, we take your privacy and data very seriously. You can review our Privacy Policy online or download the PDF.
Frequently Asked Questions
As researchers and scientists investigate more about COVID-19, different tools have emerged to detect, diagnose, and also learn more about the SARS-CoV-2 genome. Real Time RT-PCR, antibody-based methods, and antigen tests are the main tools for detecting infectious agents. The CleanPlex SARS-CoV-2 product line is intended for research use only (RUO) applications such as variant identification, mutation analysis of infectious pathogens, and not to diagnose patients for coronavirus.
