Peaks from 150 – 190 bp are residues of digested non-specific amplification products (see examples below).
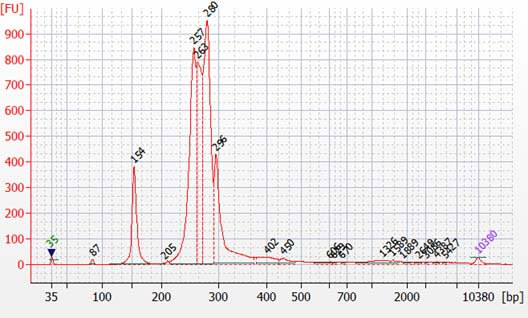
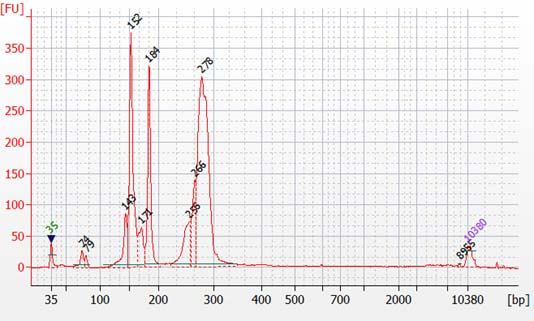
They come from incomplete removal of small-molecular-weight materials during magnetic bead purification after digestion. The digestion reagent cuts non-specific amplification products into small pieces, which are then removed during magnetic bead purification. Peaks from 150 – 190 bp are usually caused by the following:
Insufficient mixing of solutions:
Mix all working solutions and magnetic bead + sample solutions thoroughly prior to PCR and incubation steps. Vortex or pipette mix as needed to ensure solutions are homogeneous.
Inaccurate magnetic bead volume:
Again because we’re working with small volumes, it’s important to add accurate volume of beads (see 2nd page of quick guide for details). The specific bead ratio is critical for size selection in removing the smaller fragments, therefore it is crucial that the bead+sample solution is thoroughly mixed to ensure the bead to sample ratio is the same throughout.
Forgetting to add 10 uL of TE for single pool panels after mPCR:
This error is also related to bead to sample volume ratio. Add 26 uL of bead solution to 20 uL of sample ( 10ul of mPCR product + 10 uL TE) for the first purification step.
Poor Ethanol Washes:
With extremely poor ethanol washes, there can be significant carry over of byproducts to 2nd PCR step, and are then preferentially amplified. Take care to remove all supernatant after bead incubation (following the second quick spin step 5, 4, &4 on each of the purification sections), and the ethanol from the second ethanol wash of each purification step.
Extremely low or poor quality DNA input:
First, minimize freeze thaw cycles for low concentration DNA samples (<10ng/uL). Always determine the DNA concentration immediately prior to library preparation instead of days before. Higher quality samples and higher sample input tends to yield higher quality libraries with less by products.
Some custom panels might be more likely to have these products as compared to other due to the unique nature of some custom designs. If this is the case, one can perform an additional 1.3x bead to sample volume purification after the mPCR step. Or one can pool all indexed libraries (that will be sequenced in the same lane) and perform one additional round of 1.2X magnetic bead purification, before the quantification step prior to sequencing.