Rewatch the Webinar here
Access the VideoWhat is the mutation rate of SARS-CoV-2?
While it is unclear just how quickly the SARS-CoV-2 virus is mutating, we do know that there are at least 3 prominent strains in 3 different geographic regions (Asia, Europe, and North America). To stay one step ahead of potential mutations, our panel is designed with consideration of polymorphic regions of the virus and contain degenerate primers for maximum coverage.
What gene was targeted in the qPCR to determine copy number?
The primer sequences used in the qPCR referenced in the preprint were provided by CDC.
Are these the same primer sets published in the ARTIC protocol?
What was the reasoning to not include greater overlaps for the amplicons to reduce the dips in sequence coverage?
The dips in sequencing coverage are only present when using TWIST synthetic controls, which contain break points. The controls consist of six 5kb fragments, and these breakpoints are consistently present (unlike real samples) hence there is an obvious dip in which these break points are not covered. Real samples also contain break points but not all uniformly in one location. Therefore the coverage in real samples is not affected.
What is the starting material? Total RNA or RNA enriched for Viral RNA?
The input we used for clinical samples are total RNA. However enriched viral RNA and even total nucleaic acid (without DNAse) can be used as input for our assay. Our specfic primer design selectively amplify the viral genome and not the background human RNA/ DNA. We’ve also performed insilico mapping of our design against major resipiratory virues in circulation and there is negligible to no mapping to these other viral genomes
Have you compared your data to Hybrid capture based methods?
Compared to other target enrichment methods such as hybrid capture or metagenomics, amplicon-based target enrichment has shown to exhibit higher sensitivity with lower read depth requirements for the same genomic coverage, especially with low copy number inputs. Regardless of your application — strain confirmation, mutation detection, phylogenetic research, or intra-host variant calling — our amplicon-based solution achieves the highest ratios of coverage and sensitivity to sequencing read depth.
Is the reverse transcription of extracted RNA done with random hexamers?
Yes, our RT utilizes random hexamers.
For the amplicon panel, do you start with samples from saliva or nasal swabs? And are you able to consistently get enough total RNA from the extraction to go into this kit?
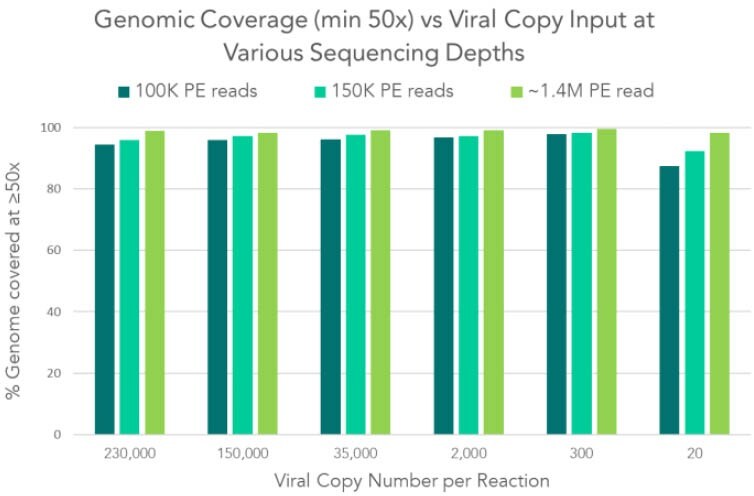
Paragon’s SARS-COV-2 panel can take RNA extracted from a various of sample types. Both saliva and nasal swab samples are compatible. Given the panel’s high sensitivity, if the extraction method yield even a few genome copies, it can be amplified and detected. We have tested clinical patient samples with down to 8 copies per reaction ( or <2 copies/uL samples).
Do you have data comparing performance among different sequencing platforms?
We’re constently gathering information with real patient sample. We have confirmed Illumina and MGI sequencing with control samples detailed in our preprint. The results are similar. The generation of quality libraries is not dependent on the sequencer to be used. Different sequencers’ performances may vary inherently due to their respective technologies.
Will libraries from negative samples be flat lines without the characteristic peaks when visualized with fragment analysis?
Negative samples may exhibit primer dimer peaks due to the lack of templates available in solution, causing a bias in dimer formation. These dimers will occur around 180 bps, which is easily distinguishable from the 280bp peak (panels for Illumina seq) from the amplified viral targets.
For Paragon, about the coverage of 99.7% are you considering on making it 100%? would it be worth it?
The <100% coverage is strictly due to amplicon-based amplification methods. The region not covered is the 92 bp in the very beginning and 92bp at the very end of the viral genome. Because amplicon-based amplification requires primer pairs for amplification, it’s not possible to design primers for a region that ends, therefore the ends of the genome are not covered. This is just a negligible fraction of the entire genome and does not impact your applications in detection, surveillance, phylogenetic studies, etc.
How sensitive is the method to RNA quality?
With 343 pairs of strategically designed primers of only 150bp in length, coupled with the already established CleanPlex Technology, this panel enables amplification of low quality RNA samples, demonstrates tolerance to viral mutations, and enables confident variant calling. CleanPlex technology has shown to perform consistently even with low quality samples (i.e. FFPE ) in other many other applications.
Is there any commercial use of the Paragon panels for diagnosis of Covid-19?
Paragon Genomics is currently selling their kits for RUO. However, we are actively pursing the right avenues to make this kit readily available to those scientists, clinicians and researchers who need it most.
Can samples be used 'directly' or is reverse transcription required as separate step?
Reverse transcription is required in order to generate the cDNA that is required for targeted amplification of SARS-CoV-2. The RT step is already included as part of the kit offerings so no need to obtain another kit for this step.
How is read depth affected in terms of amplicon based target enrichment vs hybrid capture?
Amplicon-based approach offers higher sensitivity with less sample input. Paragon’s CleanPlex technology performs on par with hybrid capture’s uniformity in coverage, but offers significantly higher coverage with fewer total reads. Compared to some hybrid capture methods on the market, our data shows more than 10x less* sequencing requirement. This enables our customers to more efficiently utilize each sequencing run to multiplex more samples.
Will other mPCR methods without digestion be able to detect mutations as well? What is the benefit of this step?
CleanPlex digestion step is used primarily to rid the final product of potential non-specific background noise that can originate from the mPCR step. This background noise is not specific to our mPCR step, however our novel approach to dealing with these off-target products is unique to the CleanPlex Technology. This allows us to generate cleaner and more robust libraries than our competition. This means with quality libraries as input, you’re more likely to obtain quality sequencing data– Quality in, Quality out.