Product Description
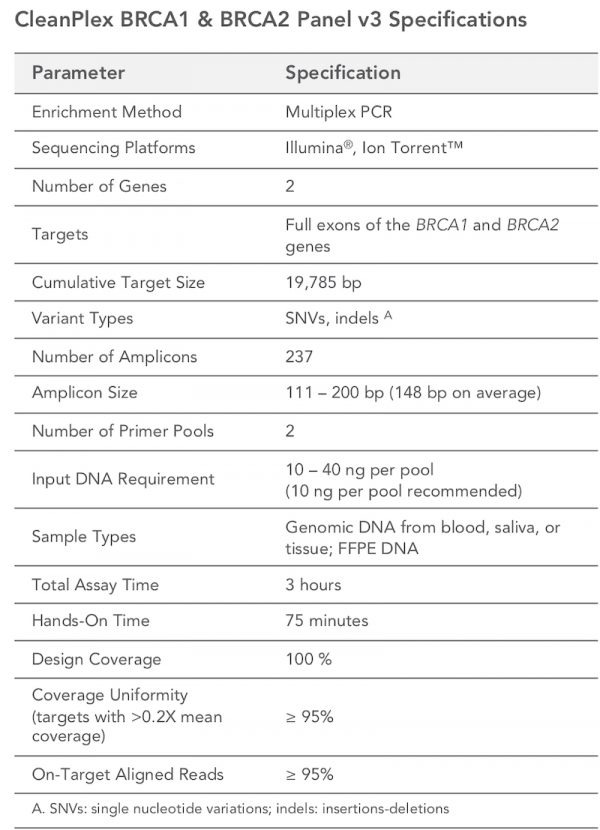
The CleanPlex BRCA1 & BRCA2 Kit v3 is a multiplex PCR- based targeted resequencing assay designed to simplify the evaluation of somatic and germline variants across BRCA1 and BRCA2 genes. The panel targets all exonic regions and 20 bp of flanking intronic sequences of BRCA1 and BRCA2. Starting with just 20 ng of DNA, sequencing-ready libraries can be prepared using a streamlined workflow in just 3 hours. The kit is optimized to deliver data with high on-target performance and high coverage uniformity to ensure efficient use of sequencing reads.
Highlights
- High Coverage of Target Regions
Target the entire coding region including 20 bases of padding around all targeted coding exons - Sensitive Detection
Detect somatic mutations as low as 1% variant allele frequency using just 20 ng of DNA - Fast, Streamlined Workflow
Generate sequencing-ready libraries in just 3 hours using a rapid, three-step protocol - Superb Performance
Prepare high-quality NGS libraries with excellent on-target performance using CleanPlex Technology to enable efficient use of sequencing reads and reduce costs
The CleanPlex BRCA1 & BRCA2 Kit v3 contains CleanPlex Multiplex PCR Primers and CleanPlex Targeted Library Kit. CleanPlex Indexed PCR Primers and CleanMag® Magnetic Beads are ordered separately to complete the workflow from input DNA to sequencing-ready NGS libraries.
Storage Temperature
Store at -20 °C.
For Research Use Only. Not for use in diagnostic procedures.
FAQs:
Question: What is BRCA?
Answer: BRCA stands for BReast CAncer. A “mutation,” or harmful genetic change, in either BRCA1 or BRCA2 gives a woman an increased lifetime risk of developing breast and ovarian cancers. Men with these gene mutations also have an increased risk of breast cancer and prostate cancer.